| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,084,651 – 7,084,756 |
| Length | 105 |
| Max. P | 0.855907 |

| Location | 7,084,651 – 7,084,756 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.49 |
| Shannon entropy | 0.59314 |
| G+C content | 0.38181 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -11.85 |
| Energy contribution | -12.96 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
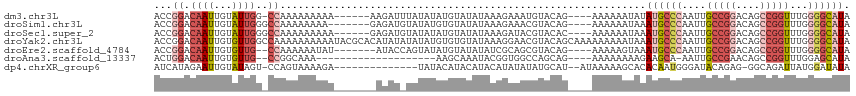
>dm3.chr3L 7084651 105 + 24543557 ACCGGACAAUUGUAUUGG-CCAAAAAAAAA------AAGAUUUAUAUAUGUAUAUAAAGAAAUGUACAG----AAAAAAUAUAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA ...((.((((...)))).-)).........------............(((((((......))))))).----........(((((((....(((((....)))))...))))))) ( -27.50, z-score = -2.79, R) >droSim1.chr3L 6590638 105 + 22553184 ACCGGACAAUUGUAUUGGGCCAAAAAAAA-------GAGAUGUAUAUGUGUAUAUAAAGAAACGUACAG----AAAAAAUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA .((((((....)).))))...........-------..........(.(((((..........))))).----)........((((((....(((((....)))))...)))))). ( -24.00, z-score = -0.92, R) >droSec1.super_2 7010961 106 + 7591821 ACCGGACAAUUGUAUUGGGCCAAAAAAAAA------GAGAUGUAUAUAUGUAUAUAAAGAUACGUACAC----AAAAAAUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA .((((((....)).))))............------....(((...(((((((......))))))).))----)........((((((....(((((....)))))...)))))). ( -26.90, z-score = -1.84, R) >droYak2.chr3L 7678191 116 + 24197627 ACCGGACAAUUGUGUUGGCCAAAAAAAAAAUACGCACAUAUAUAUAUGUGUGUAUAAAGGAACGUACAGCAAAAAAAAAUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA .((((.((((...)))).)).........(((((((((((....)))))))))))...))......................((((((....(((((....)))))...)))))). ( -37.10, z-score = -3.46, R) >droEre2.scaffold_4784 21498191 103 - 25762168 ACCGGACAAUUGUGUUG--CCAAAAAAUAU-------AUACCAGUAUAUGUAUAUAUCGCAGCGUACAG----AAAAAGUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA .........((((((((--(......((((-------((((........)))))))).)))))..))))----.........((((((....(((((....)))))...)))))). ( -29.30, z-score = -1.62, R) >droAna3.scaffold_13337 1612581 89 + 23293914 ACUGGACAAUUGUGUUG--CCGGCAAA--------------------AAGCAAAUACGGUGGCCAGCAG----AAAAAAAAGAAGCA-AAUUGCCGAACAGCCGGUUUGGAGCAUA .((((...((((((((.--...((...--------------------..)).))))))))..))))...----...........((.-....((((......)))).....))... ( -21.00, z-score = 0.21, R) >dp4.chrXR_group6 12656711 98 + 13314419 AUCAUAGAAUUGUAUAGU-CCAGUAAAAGA--------------UAUACAUACAUACAUAUAUAUGCAU--AUAAAAAGCACACAAUGGGAUACAGAG-GGCAGAUUAUGGAUAUA ..........((((((.(-(........))--------------))))))..........(((((.(((--(......((...(..((.....))..)-.))....)))).))))) ( -11.00, z-score = 0.64, R) >consensus ACCGGACAAUUGUAUUGG_CCAAAAAAAAA_______AGAU_UAUAUAUGUAUAUAAAGAAACGUACAG____AAAAAAUAAAUGCCCAAUUGCCGGACAGCCGGUUUGGGGCAUA ..................................................................................((((((....(((((....)))))...)))))). (-11.85 = -12.96 + 1.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:53 2011