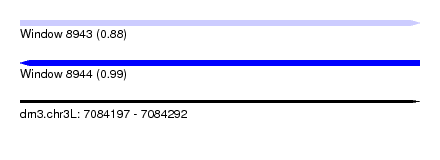
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,084,197 – 7,084,292 |
| Length | 95 |
| Max. P | 0.992793 |

| Location | 7,084,197 – 7,084,292 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 75.72 |
| Shannon entropy | 0.47374 |
| G+C content | 0.55985 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -19.24 |
| Energy contribution | -21.03 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.881846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
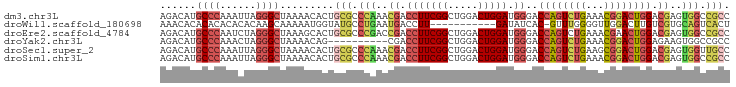
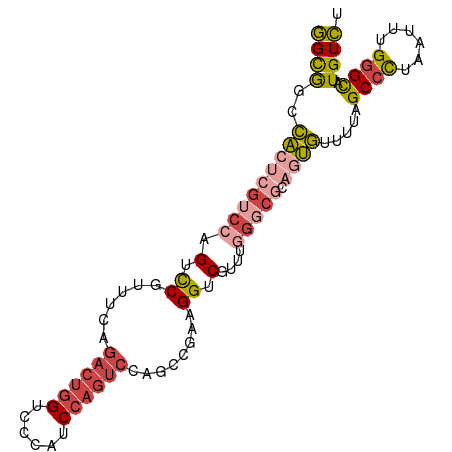
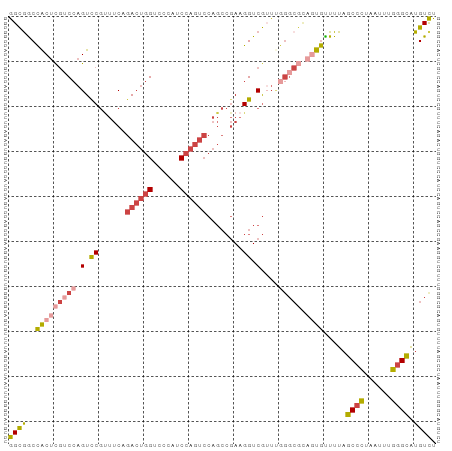
>dm3.chr3L 7084197 95 + 24543557 AGACAUGCCCAAAUUAGGGCUAAAACACUGCGCCCAAACGACCUUCGGCUGGACUGGAUGGGACCAGUCUGAAACGGACUGGACGAGUGGCCGCC ......((((......)))).........(((.(((..((.(((((((.....))))).))..((((((((...)))))))).))..))).))). ( -34.10, z-score = -2.39, R) >droWil1.scaffold_180698 1940462 83 - 11422946 AAACACACACACACACAAGCAAAAAUGGUAUGCCUGAAUGACCUU-----------GAUAUCAC-GUUUGGGGUUGGACUUGUCGUGCAGUCACU ...........(((((((((......((....)).....(((((.-----------.(......-..)..))))).).))))).)))........ ( -16.30, z-score = 1.00, R) >droEre2.scaffold_4784 21497761 95 - 25762168 AGACAUGCCCAAUCUAGGGCUAAAGCACUGCGCCCGACCGACCUUCGGCUGGACUGGAUGGGACCAGUCUGAAACGAACUGGACGAGUGGCCGCC ......((((......)))).........(((.(((..((.((((((..((((((((......))))))))...))))..)).))..))).))). ( -32.60, z-score = -1.18, R) >droYak2.chr3L 7677732 85 + 24197627 AGACAUGCCCAAACUAGGGCUAAAACAG----------CGACCUUCGGCUGGACUGGAUGGGACCAGUCUGAAACGGACUGGAGAAGUGGCCGCC ......((((......)))).......(----------((.(((((((.....))))).))..((((((((...)))))))).........))). ( -27.60, z-score = -1.15, R) >droSec1.super_2 7010497 95 + 7591821 AGACAUGCCCAAAUUAGGGCUAAAACACUGCGCCCAAACGACCUUCGGCUGGACUGGAUGGGACCAGUCUGAAGCGGACUGGACGAGUGGUUGCC ......((((......)))).........(((.(((..((.((.((.((((((((((......)))))))..))).))..)).))..))).))). ( -34.50, z-score = -2.24, R) >droSim1.chr3L 6590175 95 + 22553184 AGACAUGCCCAAAUUAGGGCUAAAACACUGCGCCCAAACGACCUUCGGCUGGACUGGAUGGGACCAGUCUGAAACGGACUGGACGAGUGGCCGCC ......((((......)))).........(((.(((..((.(((((((.....))))).))..((((((((...)))))))).))..))).))). ( -34.10, z-score = -2.39, R) >consensus AGACAUGCCCAAACUAGGGCUAAAACACUGCGCCCAAACGACCUUCGGCUGGACUGGAUGGGACCAGUCUGAAACGGACUGGACGAGUGGCCGCC ......((((......)))).........(((.(((..((.(((((((.....))))).))..((((((((...)))))))).))..))).))). (-19.24 = -21.03 + 1.80)
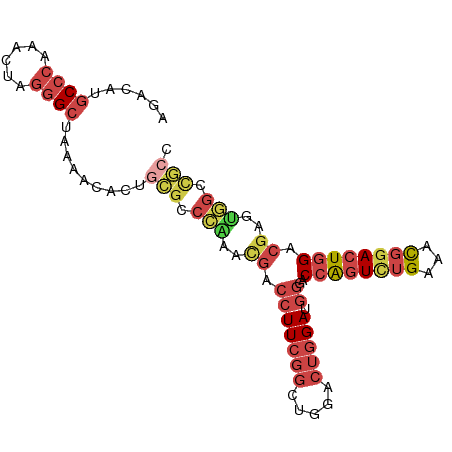
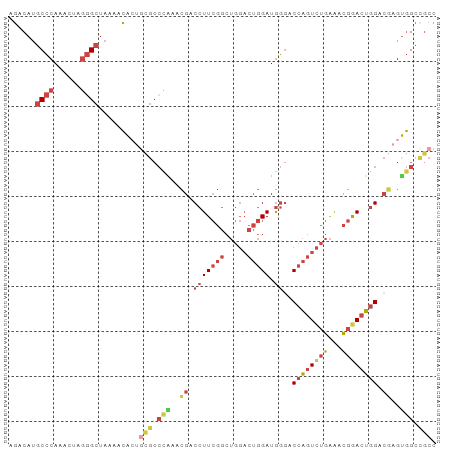
| Location | 7,084,197 – 7,084,292 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 75.72 |
| Shannon entropy | 0.47374 |
| G+C content | 0.55985 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -20.60 |
| Energy contribution | -22.65 |
| Covariance contribution | 2.05 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7084197 95 - 24543557 GGCGGCCACUCGUCCAGUCCGUUUCAGACUGGUCCCAUCCAGUCCAGCCGAAGGUCGUUUGGGCGCAGUGUUUUAGCCCUAAUUUGGGCAUGUCU .(((.(((..((.((..((.(((...((((((......)))))).))).)).)).))..))).))).........((((......))))...... ( -36.60, z-score = -2.97, R) >droWil1.scaffold_180698 1940462 83 + 11422946 AGUGACUGCACGACAAGUCCAACCCCAAAC-GUGAUAUC-----------AAGGUCAUUCAGGCAUACCAUUUUUGCUUGUGUGUGUGUGUGUUU (((((((.((((.................)-))).....-----------..)))))))((.(((((((((........))).)))))).))... ( -17.93, z-score = 0.60, R) >droEre2.scaffold_4784 21497761 95 + 25762168 GGCGGCCACUCGUCCAGUUCGUUUCAGACUGGUCCCAUCCAGUCCAGCCGAAGGUCGGUCGGGCGCAGUGCUUUAGCCCUAGAUUGGGCAUGUCU ((((((.((((((((...........((((((......))))))..((((.....)))).))))).))))))...((((......))))..))). ( -38.30, z-score = -2.24, R) >droYak2.chr3L 7677732 85 - 24197627 GGCGGCCACUUCUCCAGUCCGUUUCAGACUGGUCCCAUCCAGUCCAGCCGAAGGUCG----------CUGUUUUAGCCCUAGUUUGGGCAUGUCU (((((((.((.....))((.(((...((((((......)))))).))).)).)))))----------))......((((......))))...... ( -29.30, z-score = -2.39, R) >droSec1.super_2 7010497 95 - 7591821 GGCAACCACUCGUCCAGUCCGCUUCAGACUGGUCCCAUCCAGUCCAGCCGAAGGUCGUUUGGGCGCAGUGUUUUAGCCCUAAUUUGGGCAUGUCU ((((..(((((((((((.(((((...((((((......)))))).)))....))....))))))).)))).....((((......)))).)))). ( -37.50, z-score = -3.75, R) >droSim1.chr3L 6590175 95 - 22553184 GGCGGCCACUCGUCCAGUCCGUUUCAGACUGGUCCCAUCCAGUCCAGCCGAAGGUCGUUUGGGCGCAGUGUUUUAGCCCUAAUUUGGGCAUGUCU .(((.(((..((.((..((.(((...((((((......)))))).))).)).)).))..))).))).........((((......))))...... ( -36.60, z-score = -2.97, R) >consensus GGCGGCCACUCGUCCAGUCCGUUUCAGACUGGUCCCAUCCAGUCCAGCCGAAGGUCGUUUGGGCGCAGUGUUUUAGCCCUAAUUUGGGCAUGUCU .(((.(((..((.((..((.(((...((((((......)))))).))).)).)).))..))).))).........((((......))))...... (-20.60 = -22.65 + 2.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:53 2011