| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,057,212 – 7,057,305 |
| Length | 93 |
| Max. P | 0.702715 |

| Location | 7,057,212 – 7,057,305 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 68.37 |
| Shannon entropy | 0.56404 |
| G+C content | 0.59845 |
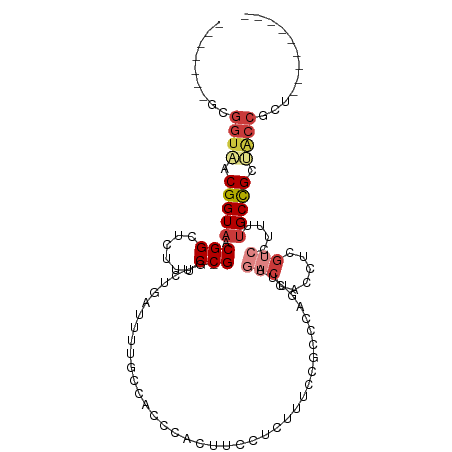
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -9.82 |
| Energy contribution | -10.99 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.702715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
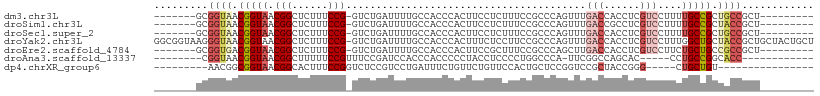
>dm3.chr3L 7057212 93 - 24543557 -------GCGGUAACGGUAACGGCUCUUUCCG-GUCUGAUUUUGCCACCCACUUCCUCUUUCCGCCCAGUUUGACCACCUCGUCCUUUUGCCGCUGCCGCU--------- -------((((((.((((((.((......))(-(((.((((..((..................))..)))).))))...........)))))).)))))).--------- ( -25.97, z-score = -2.55, R) >droSim1.chr3L 6562706 93 - 22553184 -------GCGGUAACGGUAACGGCUCUUUCCG-GUCUGAUUUUGCCACCCACUUCCUCUUUCCGCCCAGUUUGACCGCCUCGUCCUUUUGCCGCUACCGCU--------- -------((((((.((((((.((..(....((-(((.((((..((..................))..)))).)))))....)..)).)))))).)))))).--------- ( -28.27, z-score = -3.39, R) >droSec1.super_2 6983632 93 - 7591821 -------GCGGUAACGGUAACGGCUCUUUCCG-GUCUGAUUUUGCCACCCACUUCCUCUUUCCGCCCAGUUUGACCACCUCGUCCUUUUGCCGCUGCCGCU--------- -------((((((.((((((.((......))(-(((.((((..((..................))..)))).))))...........)))))).)))))).--------- ( -25.97, z-score = -2.55, R) >droYak2.chr3L 7644922 109 - 24197627 GGCGGUAAGGGUAACGGUAACGGCUCUUUCCG-GUCUGAUUUUGCCACCCACUUUCUCCUUCCGCCCAGUUUGACCACCUCGUCCUUUGGCUGCUACCGCUGCUACUGCU (((((((.((((...((((((((((......)-).)))...))))))))).............(((.((..(((.....)))..))..)))...)))))))((....)). ( -28.50, z-score = 0.19, R) >droEre2.scaffold_4784 21464097 93 + 25762168 -------GCGGUGACGGUAACGGCUCUUUCCG-GUCUGAUUUUGCCACCCACUUCCGCUUUCCGCCCAGCUUGACCACCUCGUCCUUCUGCUGCCGCCGCU--------- -------((((((.(((.(((((........(-((........)))........))).)).)))..((((..(((......))).....)))).)))))).--------- ( -24.09, z-score = -0.61, R) >droAna3.scaffold_13337 1586248 84 - 23293914 --------CGGUAACGGUAACGGCUUUUUCCGUUUCCGAUCCACCCACCCCCUACCUCCCCUGGCCCA-UUCGGCCAGCAC-----CCUGCCGGCACC------------ --------(((((.(((.(((((......))))).)))......................((((((..-...))))))...-----..))))).....------------ ( -26.60, z-score = -2.73, R) >dp4.chrXR_group6 10931641 80 + 13314419 ---------AACGGCGGUAACGGCACUUUCCGGUCUCCGUCCUGAUUUCUGUUCUGUUCCACUGCUCCGGUCCGCUACCGGG-----CUGCUGU---------------- ---------.(((((((.(((((.((..((.((.......)).)).....)).)))))))...((.(((((.....))))))-----).)))))---------------- ( -25.20, z-score = -1.12, R) >consensus _______GCGGUAACGGUAACGGCUCUUUCCG_GUCUGAUUUUGCCACCCACUUCCUCUUUCCGCCCAGUUUGACCACCUCGUCCUUUUGCCGCUACCGCU_________ .........((((.(((((..(((....((.......))....)))..........................(((......)))....))))).))))............ ( -9.82 = -10.99 + 1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:48 2011