| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,550,358 – 4,550,449 |
| Length | 91 |
| Max. P | 0.761093 |

| Location | 4,550,358 – 4,550,449 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 68.75 |
| Shannon entropy | 0.59984 |
| G+C content | 0.36093 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -6.69 |
| Energy contribution | -5.97 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
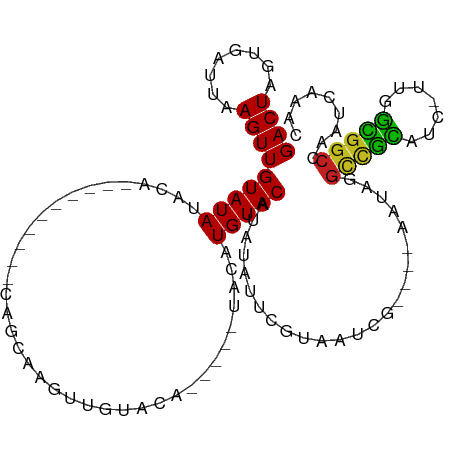
>dm3.chr2L 4550358 91 - 23011544 GACUAGUGAUUAAGUUGUAUAUA------------CAGCAAGUUGUACA-----UACCUGUACAUAUAUUCGUAAUCG----AAUAGGUCGCACC-UUGGUGGCCAAUCAAAC ......(((((..(((((....)------------))))....((((((-----....))))))..((((((....))----))))((((((...-...)))))))))))... ( -24.90, z-score = -2.48, R) >droGri2.scaffold_15126 3101239 102 + 8399593 GACUAGUCUUUAAGUUGUAUAUACAGAAAUUUGUAUAUGUGAUUAGUCA-----CAUAUGUACAUAUAUGCGUAUACA----AAUGUGCCGUGUC-UUUGCGGCCGC-CCAAC ((((((((.........(((((((((....))))))))).))))))))(-----(((.((((.((......)).))))----.))))(((((...-...)))))...-..... ( -26.10, z-score = -1.54, R) >droMoj3.scaffold_6500 11911954 102 + 32352404 GACUAGUCCUUAAGUUGUAUAUACACAAAUUUGUAUAUGUGAUUAAUCA-----CAAAUGUACAUAUAUGCGUAUACA----AAUGUGCCGUGUC-UAUGCGGCUGA-CAAAC .....(((....(((((((((.((((..((((((((((((((....)))-----)....(((......))))))))))----))).....)))).-)))))))))))-).... ( -27.50, z-score = -2.33, R) >droVir3.scaffold_12963 8461010 102 + 20206255 GACUAGUCUUUAAGUUGUAUAUACAGAAAUUUGUAUAUGUGAUUAAUCA-----CAAAUGUACAUACAUGCGUAUACA----AAUGUGCCGUGUC-UUUGCGGCCGU-CCAAC (((...........(((((((..((......(((((((((((....)))-----))..))))))....))..))))))----)..(.(((((...-...))))))))-).... ( -27.00, z-score = -2.26, R) >droWil1.scaffold_180703 1293068 111 + 3946847 GACUAGUCUUUAAGUUGUAUAUACAGAAAUUUGUAUAUGUGAUUAUCUGCCAACCAUAUGUACAUAUAUUCGUAUUCAAAACAAAAAAAAAUAAAAAAGAUGGCCGACCAA-- .....(((....(((..(((((((((....)))))))))..)))....((((...((((....))))....((.......))..................)))).)))...-- ( -16.90, z-score = -1.20, R) >droPer1.super_8 1434152 81 + 3966273 GACUAGCGAUUAAGUUGUAUAUACA----------GAAAAAGUUGUACA-----UA--UGUACAUAUUGUU-UA----------UAGGCCGCAUC-UAAGCGGCCAA---AAC ....((((((.....((((((((((----------(......)))))..-----..--)))))).))))))-..----------..((((((...-...))))))..---... ( -21.00, z-score = -2.82, R) >dp4.chr4_group3 387525 81 + 11692001 GACUAGCGAUUAAGUUGUAUAUACA----------GAAAAAGUUGUACA-----UA--UGUACAUAUUGUU-UA----------UAGGCCGCAUC-UAAGCGGCCAA---AAC ....((((((.....((((((((((----------(......)))))..-----..--)))))).))))))-..----------..((((((...-...))))))..---... ( -21.00, z-score = -2.82, R) >droAna3.scaffold_12943 1749610 98 + 5039921 GACUAGUGAUUAAGUUGUAUAUAUA------UAUGCAAAAGUUUGUACA-----UAAUUGUACAUAUACUCGUAACCG----ACUGGGCUGCAUCUUAAGCGGCCAAUCGAAC .((.((((......(((((((....------))))))).....((((((-----....))))))..)))).))...((----(...((((((.......))))))..)))... ( -26.30, z-score = -2.54, R) >droEre2.scaffold_4929 4633798 91 - 26641161 GACUAGUGAUUAAGUUGUAUAUA------------CAGCAAGUUGUACA-----UACCUGUACAUAUACUCGUAAACG----AAUAGGUCACAUC-UUGGUGGCCAAUCAAAC ......(((((..(((((....)------------))))....((((((-----....)))))).....(((....))----)...((((((...-...)))))))))))... ( -22.20, z-score = -1.64, R) >droYak2.chr2L 4567759 91 - 22324452 GACUAGUGAUUAAGUUGUAUAUA------------CAGCAAGUUGUACA-----UACAUGUACAUAUAUUCGUAAUCG----AAUAGGUCGCACC-UUGGUGGCCAAUCAAAC ......(((((..(((((....)------------))))....((((((-----....))))))..((((((....))----))))((((((...-...)))))))))))... ( -24.90, z-score = -2.59, R) >droSec1.super_5 2643101 91 - 5866729 GACUAGUGAUUAAGUUGUAUAUA------------CAGCAAGUUGUACA-----UAUCUGUACAUAUAUUCGUAAUCG----AAUAGGUCGCACC-UUGGUGGCCAAUCAAAC ......(((((..(((((....)------------))))....((((((-----....))))))..((((((....))----))))((((((...-...)))))))))))... ( -24.90, z-score = -2.68, R) >droSim1.chr2L_random 244733 91 - 909653 GACUAGUGAUUAAGUUGUAUAUA------------CAGCAAGUUGUACA-----UACCUGUACAUAUAUUCGUAAUCG----AAUAGGUCGCACC-UUGGUGGCCAAUCAAAC ......(((((..(((((....)------------))))....((((((-----....))))))..((((((....))----))))((((((...-...)))))))))))... ( -24.90, z-score = -2.48, R) >consensus GACUAGUGAUUAAGUUGUAUAUACA__________CAGCAAGUUGUACA_____UACAUGUACAUAUAUUCGUAAUCG____AAUAGGCCGCAUC_UUGGCGGCCAAUCAAAC ((((........))))(((((.....................................)))))........................(((((.......)))))......... ( -6.69 = -5.97 + -0.71)

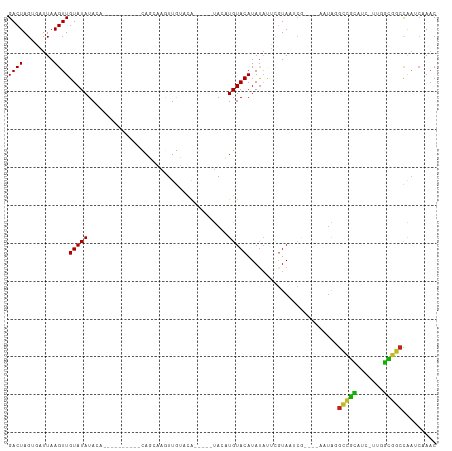
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:39 2011