
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,036,578 – 7,036,709 |
| Length | 131 |
| Max. P | 0.989985 |

| Location | 7,036,578 – 7,036,709 |
|---|---|
| Length | 131 |
| Sequences | 10 |
| Columns | 148 |
| Reading direction | forward |
| Mean pairwise identity | 62.19 |
| Shannon entropy | 0.79067 |
| G+C content | 0.68936 |
| Mean single sequence MFE | -63.79 |
| Consensus MFE | -23.81 |
| Energy contribution | -24.72 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.69 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.989985 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
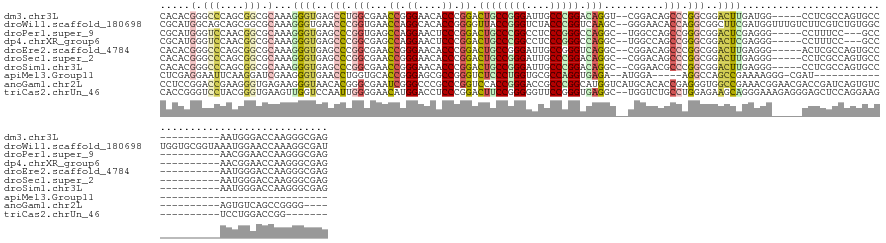
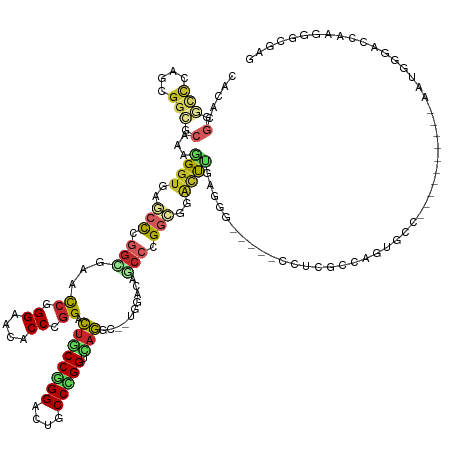
>dm3.chr3L 7036578 131 + 24543557 CACACGGGCCCAGCGGCGCAAAGGGUGAGCCUGGCGAACCGGGAACACCCGGACUGCCGGGAUUGCCCGGACAGGU--CGGACAGCCCGGCGGACUUGAUGG-----CCUCGCCAGUGCC----------AAUGGGACCAAGGGCGAG .....((.((((..(((((....((((((((...(((.(((((....))))).((((((((.(((.((((.....)--))).)))))))))))..)))..))-----.)))))).)))))----------..)))).))......... ( -66.60, z-score = -2.66, R) >droWil1.scaffold_180698 1879054 146 - 11422946 CGCAUGGCAGCAGCGGCGCAAAGGGUGAACCCGGUGAACCAGGCACACCGGGGUUACCGGGUCUACCCGGUCAAGC--GGGAACACCAGGCGGCUUCGAUGGUUUGUCUUCGUCUGUGGCUGGUGCGGUAAAUGGAACCAAAGGCGAU (((...(((.((((..(((...(((((.(((((((((.((.((....)).)).))))))))).)))))......))--)....(((.((((((...(((....)))...))))))))))))).)))(((.......)))....))).. ( -61.80, z-score = -1.25, R) >droPer1.super_9 2939907 128 - 3637205 CGCAUGGGUCCAACGGCGCAAAGGGUGAGCCCGGUGAGCCAGGAACUCCCGGACUGCCCGGCCUCCCGGGCCAGGC--UGGCCAGCCGGGCGGACUCGAGGG-----CCUUUCC---GCC----------AACGGAACCAAGGGCGAG (((.(((.(((...((((.(((((.((((.((((.(((.......))))))).((((((((((.....)))).(((--(....)))))))))).))))....-----))))).)---)))----------...))).)))...))).. ( -63.70, z-score = -0.40, R) >dp4.chrXR_group6 10910109 128 - 13314419 CGCAUGGGUCCAACGGCGCAAAGGGUGAGCCCGGCGAGCCAGGAACUCCCGGACUGCCCGGCCUCCCGGGCCAGGC--UGGCCAGCCGGGCGGACUCGAGGG-----CCUUUCC---GCC----------AACGGAACCAAGGGCGAG (((.(((.(((..((((.....(((....)))(((.((((.((.....)).....((((((....))))))..)))--).))).))))((((((((....))-----....)))---)))----------...))).)))...))).. ( -64.40, z-score = -0.41, R) >droEre2.scaffold_4784 21439539 131 - 25762168 CACACGGGCCCAGCGGCGCAAAGGGUGAGCCCGGCGAACCGGGAACACCCGGACUGCCGGGAUUGCCGGGUCAGGC--CGGACAGCCCGGCGGACUUGAGGG-----ACUCGCCAGUGCC----------AAUGGGACCAAGGGCGAG .....((.((((..(((((....(((((((((..(((.(((((....))))).((((((((.(((((((......)--))).)))))))))))..))).)))-----.)))))).)))))----------..)))).))......... ( -73.10, z-score = -3.92, R) >droSec1.super_2 6963282 131 + 7591821 CACACGGGCCCAGCGGCGCAAAGGGUGAGCCCGGCGAACCGGGAACACCCGGACUGCCGGGAUUGCCCGGACAGGC--CGGACAGCCCGGCGGACUUGAGGG-----CCUCGCCAGUGCC----------AAUGGGACCAAGGGCGAG .....((.((((..(((((....(((((((((..(((.(((((....))))).((((((((.(((.((((.....)--))).)))))))))))..))).)))-----.)))))).)))))----------..)))).))......... ( -72.80, z-score = -3.67, R) >droSim1.chr3L 6542193 131 + 22553184 CACACGGGCCCAGCGGCGCAAAGGGUGAGCCCGGCGAACCGGGAACACCCGGACUGCCGGGAUUGCCCGGACAGGC--CGGAACGCCCGGCGGACUUGAGGG-----CCUCGCCAGUGCC----------AAUGGGACCAAGGGCGAG .....((.((((..(((((...(((((...(((((...(((((....))))).((((((((....))))).)))))--)))..)))))(((((.((....))-----.).)))).)))))----------..)))).))......... ( -70.30, z-score = -2.96, R) >apiMel3.Group11 5560736 101 - 12576330 CUCGAGGAAUUCAAGGAUCGAAGGGUGAACCUGGUGCACCGGGAGCGCCGGGUCUCCCUGGUGCGCCAGGUGAGA--AUGGA-----AGGCCAGCCGAAAAGGG-CGAU--------------------------------------- .....((..(((....(((....)))..((((((((((((((((((.....).))))).))))))))))))....--...))-----)..)).(((......))-)...--------------------------------------- ( -46.60, z-score = -2.95, R) >anoGam1.chr2L 33150669 134 + 48795086 CCUCCGGACCGAAGGGUGAGAAGGGUAACACGGGCGAAUCGGGCCCGCCCGGUCCACCGGGACCGCCCGGCAUGGUCAUGCACACCGAGGGUGGCCGAAACGGAACGACCGAUCAGUGUC----------AGUGUCAGCCGGGG---- .(((((((((....)))..........((((((((.....(((....)))(((((....)))))))))(((((((((((.(.......).))))))....(((.....)))....)))))----------.))))...))))))---- ( -59.00, z-score = 0.02, R) >triCas2.chrUn_46 154451 129 - 315570 CACCGGGUCCUACGGGUGAAGUUGGUCCAAUUGGGGAACAUGGACCUCCCGGACUUCCGGGGGUUCCGGGUGAGGC--UGGUCUGCCUGGAGAAGCAGGGAAAGAGGGAGCUCCAGGAAG----------UCCUGGACCGG------- ..((((.(((...(((.......((((((....(....).)))))).)))((((((((.((((((((.((.....)--)..(((.((((......))))...))).)))))))).)))))----------))).)))))))------- ( -59.60, z-score = -1.03, R) >consensus CACACGGGCCCAGCGGCGCAAAGGGUGAGCCCGGCGAACCGGGAACACCCGGACUGCCGGGACUGCCCGGUCAGGC__UGGACAGCCCGGCGGACUUGAGGG_____CCUCGCCAGUGCC__________AAUGGGACCAAGGGCGAG .....(.(((....))).)...((((..(((.(((...((.((....)).)).((((((((....))))).)))..........))).)))..))))................................................... (-23.81 = -24.72 + 0.91)
| Location | 7,036,578 – 7,036,709 |
|---|---|
| Length | 131 |
| Sequences | 10 |
| Columns | 148 |
| Reading direction | reverse |
| Mean pairwise identity | 62.19 |
| Shannon entropy | 0.79067 |
| G+C content | 0.68936 |
| Mean single sequence MFE | -60.76 |
| Consensus MFE | -19.88 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981115 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
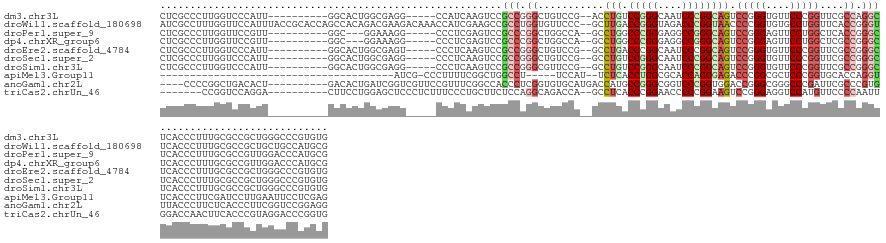
>dm3.chr3L 7036578 131 - 24543557 CUCGCCCUUGGUCCCAUU----------GGCACUGGCGAGG-----CCAUCAAGUCCGCCGGGCUGUCCG--ACCUGUCCGGGCAAUCCCGGCAGUCCGGGUGUUCCCGGUUCGCCAGGCUCACCCUUUGCGCCGCUGGGCCCGUGUG ..(((...(((.((((.(----------(((.((((((((.-----...........((((((.((((((--.......))))))..))))))...(((((....)))))))))))))((.........)))))).)))).))).))) ( -63.80, z-score = -2.15, R) >droWil1.scaffold_180698 1879054 146 + 11422946 AUCGCCUUUGGUUCCAUUUACCGCACCAGCCACAGACGAAGACAAACCAUCGAAGCCGCCUGGUGUUCCC--GCUUGACCGGGUAGACCCGGUAACCCCGGUGUGCCUGGUUCACCGGGUUCACCCUUUGCGCCGCUGCUGCCAUGCG ..(((....(((.......)))(((.((((......(((.(......).)))..(.(((..((((..(((--(...(((((((((.(((.((....)).))).)))))))))...))))..))))....))).))))).)))...))) ( -55.20, z-score = -2.27, R) >droPer1.super_9 2939907 128 + 3637205 CUCGCCCUUGGUUCCGUU----------GGC---GGAAAGG-----CCCUCGAGUCCGCCCGGCUGGCCA--GCCUGGCCCGGGAGGCCGGGCAGUCCGGGAGUUCCUGGCUCACCGGGCUCACCCUUUGCGCCGUUGGACCCAUGCG ..(((...(((.((((.(----------(((---(.(((((-----.....((((((((((((((((((.--....)))).....))))))))((.(((((....)))))))....))))))..))))).))))).)))).))).))) ( -70.20, z-score = -2.02, R) >dp4.chrXR_group6 10910109 128 + 13314419 CUCGCCCUUGGUUCCGUU----------GGC---GGAAAGG-----CCCUCGAGUCCGCCCGGCUGGCCA--GCCUGGCCCGGGAGGCCGGGCAGUCCGGGAGUUCCUGGCUCGCCGGGCUCACCCUUUGCGCCGUUGGACCCAUGCG ..(((...(((.((((.(----------(((---(.(((((-----.......((..(((((((.((((.--((((((((.....)))))))).....(((....))))))).)))))))..))))))).))))).)))).))).))) ( -70.91, z-score = -1.83, R) >droEre2.scaffold_4784 21439539 131 + 25762168 CUCGCCCUUGGUCCCAUU----------GGCACUGGCGAGU-----CCCUCAAGUCCGCCGGGCUGUCCG--GCCUGACCCGGCAAUCCCGGCAGUCCGGGUGUUCCCGGUUCGCCGGGCUCACCCUUUGCGCCGCUGGGCCCGUGUG ..(((...(((.((((.(----------(((...((((.(.-----(......).)))))((((((.(((--(((......))).....)))))))))(((((..(((((....)))))..))))).....)))).)))).))).))) ( -62.80, z-score = -1.77, R) >droSec1.super_2 6963282 131 - 7591821 CUCGCCCUUGGUCCCAUU----------GGCACUGGCGAGG-----CCCUCAAGUCCGCCGGGCUGUCCG--GCCUGUCCGGGCAAUCCCGGCAGUCCGGGUGUUCCCGGUUCGCCGGGCUCACCCUUUGCGCCGCUGGGCCCGUGUG ..(((...(((.((((.(----------(((.((((((.((-----(......)))))))))((.....(--(.(((.(((((....)))))))).))(((((..(((((....)))))..)))))...)))))).)))).))).))) ( -70.20, z-score = -2.81, R) >droSim1.chr3L 6542193 131 - 22553184 CUCGCCCUUGGUCCCAUU----------GGCACUGGCGAGG-----CCCUCAAGUCCGCCGGGCGUUCCG--GCCUGUCCGGGCAAUCCCGGCAGUCCGGGUGUUCCCGGUUCGCCGGGCUCACCCUUUGCGCCGCUGGGCCCGUGUG ..(((...(((.((((.(----------(((.((((((.((-----(......)))))))))(((....(--(.(((.(((((....)))))))).))(((((..(((((....)))))..)))))..))))))).)))).))).))) ( -70.70, z-score = -2.98, R) >apiMel3.Group11 5560736 101 + 12576330 ---------------------------------------AUCG-CCCUUUUCGGCUGGCCU-----UCCAU--UCUCACCUGGCGCACCAGGGAGACCCGGCGCUCCCGGUGCACCAGGUUCACCCUUCGAUCCUUGAAUUCCUCGAG ---------------------------------------((((-((......)))(((...-----.))).--....((((((.(((((.(((((........)))))))))).)))))).........))).(((((.....))))) ( -36.40, z-score = -2.21, R) >anoGam1.chr2L 33150669 134 - 48795086 ----CCCCGGCUGACACU----------GACACUGAUCGGUCGUUCCGUUUCGGCCACCCUCGGUGUGCAUGACCAUGCCGGGCGGUCCCGGUGGACCGGGCGGGCCCGAUUCGCCCGUGUUACCCUUCUCACCCUUCGGUCCGGAGG ----((((((((((...(----------(((((((...(((((........)))))((((((((((((......))))))))).)))..)))))(((((((((((.....)))))))).))).......)))....)))).)))).)) ( -57.30, z-score = -0.02, R) >triCas2.chrUn_46 154451 129 + 315570 -------CCGGUCCAGGA----------CUUCCUGGAGCUCCCUCUUUCCCUGCUUCUCCAGGCAGACCA--GCCUCACCCGGAACCCCCGGAAGUCCGGGAGGUCCAUGUUCCCCAAUUGGACCAACUUCACCCGUAGGACCCGGUG -------((((((((((.----------...))))))............(((((......((((......--)))).....((....(((((....))))).((((((...........))))))........)))))))..)))).. ( -50.10, z-score = -1.79, R) >consensus CUCGCCCUUGGUCCCAUU__________GGCACUGGCGAGG_____CCCUCAAGUCCGCCGGGCUGUCCA__GCCUGACCGGGCAAUCCCGGCAGUCCGGGUGUUCCCGGUUCGCCGGGCUCACCCUUUGCGCCGCUGGGCCCGUGUG .........................................................(((.((...........(((.(((((....)))))))).(((((....)))))....)).)))............................ (-19.88 = -20.48 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:45 2011