| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,030,952 – 7,031,073 |
| Length | 121 |
| Max. P | 0.968576 |

| Location | 7,030,952 – 7,031,073 |
|---|---|
| Length | 121 |
| Sequences | 7 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Shannon entropy | 0.43838 |
| G+C content | 0.51088 |
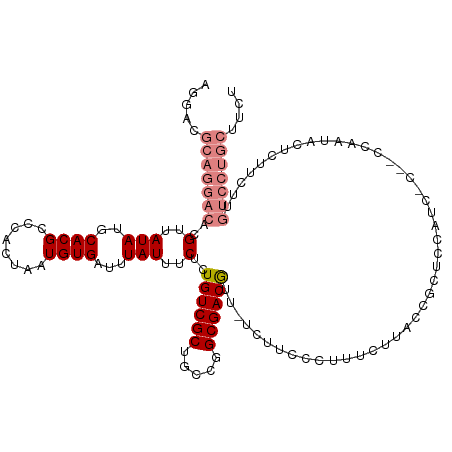
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -17.54 |
| Energy contribution | -19.41 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
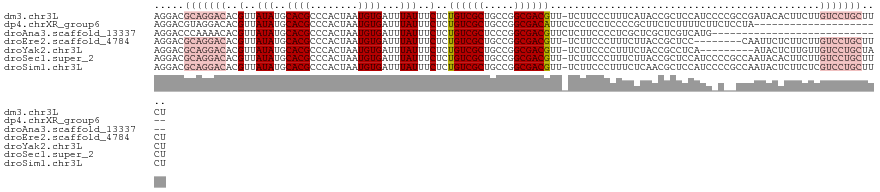
>dm3.chr3L 7030952 121 + 24543557 AGGACGCAGGACACGUUAUAUGCACGCCCACUAAUGUGAUUUAUUUCUCUGUCGCUGCCGGCGACGUU-UCUUCCCUUUCAUACCGCUCCAUCCCCGCCGAUACACUUCUUGUCCUGCUUCU .(((.((((((((........((.....(((....)))............(((((.....)))))...-................))............((......)).)))))))).))) ( -28.40, z-score = -1.84, R) >dp4.chrXR_group6 10903968 100 - 13314419 AGGACGUAGGACACGUUAUAUGCACGCCCACUAAUGUGAUUUAUUUCUCUGUCGCUGCCGGCGACAUUCUCCUCCUCCCCGCUUCUCUUUUCUUCUCCUA---------------------- .(((.(.(((((((((((.............)))))))...........((((((.....))))))...))))).)))......................---------------------- ( -20.82, z-score = -1.63, R) >droAna3.scaffold_13337 1560743 93 + 23293914 AGGACCCAAAACACGUUAUAUGCACGCCCACUAAUGUGAUUUAUUUCUCUGUCGCUCCCGGCGACGUUCUCUUCCCCUCGCUCGCUCGUCAUG----------------------------- (((((......(((((((.............)))))))............(((((.....)))))))))).......................----------------------------- ( -15.92, z-score = -0.29, R) >droEre2.scaffold_4784 21433884 113 - 25762168 AGGACGCAGGACACGUUAUAUGCACGCCCACUAAUGUGAUUUAUUUCUCUGUCGCUGCCGGCGACGUU-UCUUCCCUUUCUUACCGCUCC--------CAAUUCUCUUCUUGUCCUGCUUCU .(((.((((((((.((.....))..((.(((....)))............(((((.....)))))...-................))...--------............)))))))).))) ( -27.90, z-score = -2.88, R) >droYak2.chr3L 7616579 112 + 24197627 AGGACGCAGGACACGUUAUAUGCACGCCCACUAAUGUGAUUUAUUUCUCUGUCGCUGCCGGCGACGUU-UCUUCCCCUUUCUACCGCCUCA---------AUACUCUUGUUGUCCUGCUACU .....((((((((((...(((....((.(((....)))............(((((.....)))))...-................))....---------)))....)).)))))))).... ( -26.30, z-score = -1.73, R) >droSec1.super_2 6957597 121 + 7591821 AGGACGCAGGACACGUUAUAUGCACGCCCACUAAUGUGAUUUAUUUCUCUGUCGCUGCCGGCGACGUU-UCUUCCCUUUCUUACCGCUCCAUCCCCGCCAAUACACUUCUUGUCCUGCUUCU .(((.((((((((.((.....))..((.(((....)))............(((((.....)))))...-................)).......................)))))))).))) ( -27.90, z-score = -2.26, R) >droSim1.chr3L 6536504 121 + 22553184 AGGACGCAGGACACGUUAUAUGCACGCCCACUAAUGUGAUUUAUUUCUCUGUCGCUGCCGGCGACGUU-UCUUCCCUUUCUCAACGCUCCAUCCCCGCCAAUACUCUUCUCGUCCUGCUUCU .(((.(((((((..((.....))..((.(((....)))............(((((.....)))))...-................))........................))))))).))) ( -27.60, z-score = -2.06, R) >consensus AGGACGCAGGACACGUUAUAUGCACGCCCACUAAUGUGAUUUAUUUCUCUGUCGCUGCCGGCGACGUU_UCUUCCCUUUCUUACCGCUCCAUC_C__CCAAUACUCUUCUUGUCCUGCUUCU .....(((((((..(..(((..((((........))))...)))..)..((((((.....)))))).............................................))))))).... (-17.54 = -19.41 + 1.88)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:44 2011