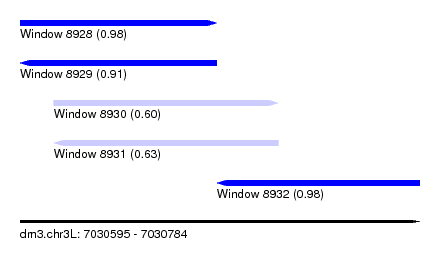
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 7,030,595 – 7,030,784 |
| Length | 189 |
| Max. P | 0.981381 |

| Location | 7,030,595 – 7,030,688 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.55 |
| Shannon entropy | 0.47589 |
| G+C content | 0.43794 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -13.71 |
| Energy contribution | -14.05 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
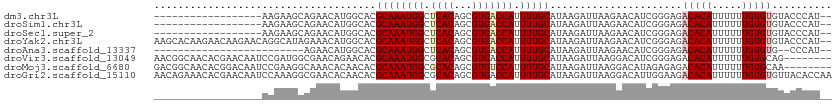
>dm3.chr3L 7030595 93 + 24543557 ------------------AAGAAGCAGAACAUGGCACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCGGGAGACACAUUUUUUGUGUGUACCCAU-- ------------------.....(((((..(((((((((...((.....))))))).))))))))).................((((.(((((.....))))).).)))..-- ( -28.60, z-score = -3.54, R) >droSim1.chr3L 6536147 93 + 22553184 ------------------AAGAAGCAGAACAUGGCACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCGGGAGACACAUUUUUUGUGUGUACCCAU-- ------------------.....(((((..(((((((((...((.....))))))).))))))))).................((((.(((((.....))))).).)))..-- ( -28.60, z-score = -3.54, R) >droSec1.super_2 6957262 93 + 7591821 ------------------AAGAAGCAGAACAUGGCACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCGGGAGACACAUUUUUUGUGUGUACCCAU-- ------------------.....(((((..(((((((((...((.....))))))).))))))))).................((((.(((((.....))))).).)))..-- ( -28.60, z-score = -3.54, R) >droYak2.chr3L 7616202 111 + 24197627 AAGCACAAGAACAAGAACAGGCAUAGAAACAUGGCACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCGGGAGACACAUUUUUUGUGUGUACCCAU-- ..............((...(.(((......))).)..((((((((.((((...))))))).)))))...............))((((.(((((.....))))).).)))..-- ( -25.50, z-score = -1.63, R) >droAna3.scaffold_13337 1560412 84 + 23293914 -------------------------AGAACAUGGCACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCGGGAGACACAUUUUUUGUGUG--CCCAU-- -------------------------.......(((((((((((((.((((...))))))).))))).......((((((.....(....)....)))))).)))--))...-- ( -22.60, z-score = -1.80, R) >droVir3.scaffold_13049 7481881 105 + 25233164 AACGGCAACACGAACAAUCCGAUGGCGAACAGAACACGCAAAUGGCGCACAGCGUGACCAUUUUGCAUAAGAUUAAGGACAUCGGGAGACACAUUUUUUGUGCAG-------- ...(....)........((((((((((.........)))(((((((((.....))).))))))................))))))).(.((((.....)))))..-------- ( -25.10, z-score = -1.66, R) >droMoj3.scaffold_6680 5732934 105 + 24764193 GACGGCAACACGGACAAUCCGAAGGCAAACACAACACGCAAAUGGCGCACAGCGUGUCCAUUUUGCAUAAGAUUAAGGACAUAGAGAGACACAUUUUUUGUGCAA-------- ...(....).(((.....)))...(((((....((((((...((.....))))))))....)))))...........(.(((((((((.....))))))))))..-------- ( -22.30, z-score = -1.00, R) >droGri2.scaffold_15110 19166121 113 - 24565398 AACAGAAACACGAACAAUCCAAAGGCGAACACAACACGCAAAUGGCGCACAGCGUGACCAUUUUGCAUAAGAUUAAGGACAUUGGAAGACACAUUUUUUGUGUGUUACACCAA .................(((...(.....).......(((((((((((.....))).))).)))))..........)))..((((..(((((((.....)))))))...)))) ( -23.00, z-score = -1.05, R) >consensus __________________AAGAAGCAGAACAUGGCACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCGGGAGACACAUUUUUUGUGUGUACCCAU__ .....................................((((((((.((((...))))))).)))))......................(((((.....))))).......... (-13.71 = -14.05 + 0.34)

| Location | 7,030,595 – 7,030,688 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.55 |
| Shannon entropy | 0.47589 |
| G+C content | 0.43794 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -15.65 |
| Energy contribution | -15.59 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.908683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7030595 93 - 24543557 --AUGGGUACACACAAAAAAUGUGUCUCCCGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUGCCAUGUUCUGCUUCUU------------------ --..(((...(((((.....)))))..)))(((.......)))....(((..((((.(((((.(((...)))..)))))))))....))).....------------------ ( -25.20, z-score = -2.23, R) >droSim1.chr3L 6536147 93 - 22553184 --AUGGGUACACACAAAAAAUGUGUCUCCCGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUGCCAUGUUCUGCUUCUU------------------ --..(((...(((((.....)))))..)))(((.......)))....(((..((((.(((((.(((...)))..)))))))))....))).....------------------ ( -25.20, z-score = -2.23, R) >droSec1.super_2 6957262 93 - 7591821 --AUGGGUACACACAAAAAAUGUGUCUCCCGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUGCCAUGUUCUGCUUCUU------------------ --..(((...(((((.....)))))..)))(((.......)))....(((..((((.(((((.(((...)))..)))))))))....))).....------------------ ( -25.20, z-score = -2.23, R) >droYak2.chr3L 7616202 111 - 24197627 --AUGGGUACACACAAAAAAUGUGUCUCCCGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUGCCAUGUUUCUAUGCCUGUUCUUGUUCUUGUGCUU --..(((...(((((.....)))))..)))(((.......)))....(((..((((.(((((.(((...)))..)))))))))((......))...............))).. ( -23.60, z-score = -0.45, R) >droAna3.scaffold_13337 1560412 84 - 23293914 --AUGGG--CACACAAAAAAUGUGUCUCCCGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUGCCAUGUUCU------------------------- --..(((--.(((((.....)))))..)))(((.......))).((((((((.(((((((...)))).))))))))))).........------------------------- ( -22.50, z-score = -1.57, R) >droVir3.scaffold_13049 7481881 105 - 25233164 --------CUGCACAAAAAAUGUGUCUCCCGAUGUCCUUAAUCUUAUGCAAAAUGGUCACGCUGUGCGCCAUUUGCGUGUUCUGUUCGCCAUCGGAUUGUUCGUGUUGCCGUU --------..(((((.....)))))...((((((..........((((((((.(((((((...))).))))))))))))..........)))))).................. ( -25.45, z-score = -1.10, R) >droMoj3.scaffold_6680 5732934 105 - 24764193 --------UUGCACAAAAAAUGUGUCUCUCUAUGUCCUUAAUCUUAUGCAAAAUGGACACGCUGUGCGCCAUUUGCGUGUUGUGUUUGCCUUCGGAUUGUCCGUGUUGCCGUC --------..(((((.....)))))......................((((.((((((((((.(..(((.....)))..).))))....(....)...)))))).)))).... ( -26.70, z-score = -1.86, R) >droGri2.scaffold_15110 19166121 113 + 24565398 UUGGUGUAACACACAAAAAAUGUGUCUUCCAAUGUCCUUAAUCUUAUGCAAAAUGGUCACGCUGUGCGCCAUUUGCGUGUUGUGUUCGCCUUUGGAUUGUUCGUGUUUCUGUU ..((((.((((((((.....))).....................((((((((.(((((((...))).))))))))))))..)))))))))....................... ( -23.90, z-score = -0.21, R) >consensus __AUGGGUACACACAAAAAAUGUGUCUCCCGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUGCCAUGUUCUCCUUCGU__________________ ..........(((((.....)))))...................((((((((.(((((((...))).)))))))))))).................................. (-15.65 = -15.59 + -0.06)

| Location | 7,030,611 – 7,030,717 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.29 |
| Shannon entropy | 0.49782 |
| G+C content | 0.44211 |
| Mean single sequence MFE | -24.13 |
| Consensus MFE | -12.59 |
| Energy contribution | -12.88 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.598064 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7030611 106 + 24543557 CACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCG-GGAGACACAUUUUUUGUGUGUA---CCCAUCCAUGUAGC-----CUAUUGGAAUGUCGGUAGAC- ...((((((((.((((...))))))))))(((((..(((.......)))(-(((.(((((.....))))).).---)))....)))))))-----(((((((....)))))))..- ( -28.10, z-score = -1.18, R) >droSim1.chr3L 6536163 107 + 22553184 CACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCG-GGAGACACAUUUUUUGUGUGUA---CCCAUCCAUGUAGC-----CUAUUGGAAUGUCGGUGGACC ...((((((((.((((...))))))).))))).............(((((-(((.(((((.....))))).).---))).((((......-----....)))).....)))).... ( -30.30, z-score = -1.26, R) >droSec1.super_2 6957278 85 + 7591821 CACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCG-GGAGACACAUUUUUUGUGUGUA---CCCAUCCAUGUAC--------------------------- ...((((((((.((((...))))))).))))).................(-(((.(((((.....))))).).---)))..........--------------------------- ( -23.10, z-score = -2.21, R) >droYak2.chr3L 7616236 106 + 24197627 CACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCG-GGAGACACAUUUUUUGUGUGUA---CCCAUCCAUGCAGC-----CUAUUGGAAUGUCGGUAGAC- ...((((((((.((((...))))))))))(((((..(((.......)))(-(((.(((((.....))))).).---)))....)))))))-----(((((((....)))))))..- ( -30.10, z-score = -1.75, R) >droEre2.scaffold_4784 21433526 103 - 25762168 CACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCG-G-AGACACAUUUUUUGUGUGUA---CCCAUCCAUGUAGC-----CUAUUGGAAUGUCGGUAG--- ...((((((((.((((...))))))).)))))............((((.(-(-(.((((((.....)))))).---....)))))))...-----(((((((....)))))))--- ( -26.50, z-score = -1.23, R) >droAna3.scaffold_13337 1560421 95 + 23293914 CACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCG-GGAGACACAUUUUUUGUGUG-----CCCAUCCAUGUCGG------UAUUGGAAUGC--------- ...((((((((.((((...))))))).))))).................(-((..(((((.....))))).-----))).((((......------...))))....--------- ( -25.60, z-score = -0.98, R) >dp4.chrXR_group6 10903589 100 - 13314419 CACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUUC-GGAGACACAUUUUUUGUGUA-----CCCAUCCAUGUCGCUGCCGCUAUUGGAAUG---------- ...((((((((.((((...))))))).)))))..................-((..(((((.....))))).-----.)).((((....((....))...))))...---------- ( -24.90, z-score = -1.17, R) >droPer1.super_9 2933320 100 - 3637205 CACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUUC-GGAGACACAUUUUUUGUGUA-----CCCAUCCAUGUCGCUGCCGCUAUUGGAAUG---------- ...((((((((.((((...))))))).)))))..................-((..(((((.....))))).-----.)).((((....((....))...))))...---------- ( -24.90, z-score = -1.17, R) >droWil1.scaffold_180698 1870017 82 - 11422946 CACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUUUUGGAGACACAUUUUUUUCCAUGU---AAAGACCAC------------------------------- ...((((((((.((((...))))))).)))))............(((...((((((........)))))))))---.........------------------------------- ( -16.30, z-score = -1.95, R) >droVir3.scaffold_13049 7481915 92 + 25233164 CACGCAAAUGGCGCACAGCGUGACCAUUUUGCAUAAGAUUAAGGACAUCG-GGAGACACAUUUUUUGUGCAGC----CAGACCACAAACACAAAGCU------------------- ...(((((((((((.....))).))).)))))..........((...((.-((...((((.....))))...)----).))))..............------------------- ( -19.20, z-score = -0.73, R) >droMoj3.scaffold_6680 5732968 96 + 24764193 CACGCAAAUGGCGCACAGCGUGUCCAUUUUGCAUAAGAUUAAGGACAUAG-AGAGACACAUUUUUUGUGCAACACCACAGCCAACGUACCCACAGCU------------------- ...((...((((.......(((((((((((....)))))...))))))..-...(.((((.....))))).........))))..((....)).)).------------------- ( -17.90, z-score = -0.16, R) >droGri2.scaffold_15110 19166155 105 - 24565398 CACGCAAAUGGCGCACAGCGUGACCAUUUUGCAUAAGAUUAAGGACAUUG-GAAGACACAUUUUUUGUGUGUUACACCAACCAACACCAACAAUAUACAAAAAGCA---------- ...(((((((((((.....))).))).)))))..........((...(((-(..(((((((.....)))))))...))))))........................---------- ( -22.70, z-score = -1.67, R) >consensus CACGCAAAUGGCUCACAGCGUGACCAUUUUGCAUAAGAUUAAGAACAUCG_GGAGACACAUUUUUUGUGUGUA___CCCAUCCAUGUAGC_____CUAUUGGAAUG__________ ...((((((((.((((...))))))).)))))........................((((.....))))............................................... (-12.59 = -12.88 + 0.29)

| Location | 7,030,611 – 7,030,717 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.29 |
| Shannon entropy | 0.49782 |
| G+C content | 0.44211 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -14.95 |
| Energy contribution | -14.93 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7030611 106 - 24543557 -GUCUACCGACAUUCCAAUAG-----GCUACAUGGAUGGG---UACACACAAAAAAUGUGUCUCC-CGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUG -(((....))).......(((-----(..((((...((((---...(((((.....)))))..))-)))))).))))....((((((((.(((((((...)))).))))))))))) ( -27.70, z-score = -1.28, R) >droSim1.chr3L 6536163 107 - 22553184 GGUCCACCGACAUUCCAAUAG-----GCUACAUGGAUGGG---UACACACAAAAAAUGUGUCUCC-CGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUG ((....))(((((((((....-----......))))((((---...(((((.....)))))..))-)))))))........((((((((.(((((((...)))).))))))))))) ( -27.70, z-score = -0.67, R) >droSec1.super_2 6957278 85 - 7591821 ---------------------------GUACAUGGAUGGG---UACACACAAAAAAUGUGUCUCC-CGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUG ---------------------------(.((((...((((---...(((((.....)))))..))-)))))).).......((((((((.(((((((...)))).))))))))))) ( -23.20, z-score = -1.52, R) >droYak2.chr3L 7616236 106 - 24197627 -GUCUACCGACAUUCCAAUAG-----GCUGCAUGGAUGGG---UACACACAAAAAAUGUGUCUCC-CGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUG -(((....))).......((.-----(((((((((..(((---...(((((.....)))))..))-)(((.......))))))))))((((((((((...)))).)))))))).)) ( -28.30, z-score = -1.12, R) >droEre2.scaffold_4784 21433526 103 + 25762168 ---CUACCGACAUUCCAAUAG-----GCUACAUGGAUGGG---UACACACAAAAAAUGUGUCU-C-CGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUG ---.....(((((((((....-----......))))(((.---.(((((.......)))))..-)-)))))))........((((((((.(((((((...)))).))))))))))) ( -24.80, z-score = -0.73, R) >droAna3.scaffold_13337 1560421 95 - 23293914 ---------GCAUUCCAAUA------CCGACAUGGAUGGG-----CACACAAAAAAUGUGUCUCC-CGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUG ---------...........------..(((((.(..(((-----((((.......)))))))..-).)))))........((((((((.(((((((...)))).))))))))))) ( -25.70, z-score = -1.33, R) >dp4.chrXR_group6 10903589 100 + 13314419 ----------CAUUCCAAUAGCGGCAGCGACAUGGAUGGG-----UACACAAAAAAUGUGUCUCC-GAAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUG ----------...((((...((....))....))))(((.-----.(((((.....)))))..))-)..............((((((((.(((((((...)))).))))))))))) ( -27.20, z-score = -0.96, R) >droPer1.super_9 2933320 100 + 3637205 ----------CAUUCCAAUAGCGGCAGCGACAUGGAUGGG-----UACACAAAAAAUGUGUCUCC-GAAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUG ----------...((((...((....))....))))(((.-----.(((((.....)))))..))-)..............((((((((.(((((((...)))).))))))))))) ( -27.20, z-score = -0.96, R) >droWil1.scaffold_180698 1870017 82 + 11422946 -------------------------------GUGGUCUUU---ACAUGGAAAAAAAUGUGUCUCCAAAAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUG -------------------------------.(((....(---((((........)))))...)))...............((((((((.(((((((...)))).))))))))))) ( -19.00, z-score = -2.07, R) >droVir3.scaffold_13049 7481915 92 - 25233164 -------------------AGCUUUGUGUUUGUGGUCUG----GCUGCACAAAAAAUGUGUCUCC-CGAUGUCCUUAAUCUUAUGCAAAAUGGUCACGCUGUGCGCCAUUUGCGUG -------------------...............(((.(----(..(((((.....)))))..))-.)))...........((((((((.(((((((...))).)))))))))))) ( -21.60, z-score = -0.44, R) >droMoj3.scaffold_6680 5732968 96 - 24764193 -------------------AGCUGUGGGUACGUUGGCUGUGGUGUUGCACAAAAAAUGUGUCUCU-CUAUGUCCUUAAUCUUAUGCAAAAUGGACACGCUGUGCGCCAUUUGCGUG -------------------.((.((((((((...(((.((((.(..(((((.....)))))..).-))))((((.................))))..))))))).))))..))... ( -26.53, z-score = -0.86, R) >droGri2.scaffold_15110 19166155 105 + 24565398 ----------UGCUUUUUGUAUAUUGUUGGUGUUGGUUGGUGUAACACACAAAAAAUGUGUCUUC-CAAUGUCCUUAAUCUUAUGCAAAAUGGUCACGCUGUGCGCCAUUUGCGUG ----------.((((((((((((..(((((.(...(((((....(((((.......)))))...)-))))..).)))))..))))))))).)))(((((.(((...)))..))))) ( -24.70, z-score = -0.63, R) >consensus __________CAUUCCAAUAG_____GCUACAUGGAUGGG___UACACACAAAAAAUGUGUCUCC_CGAUGUUCUUAAUCUUAUGCAAAAUGGUCACGCUGUGAGCCAUUUGCGUG .................................((((.........(((((.....))))).........)))).......((((((((.(((((((...))).)))))))))))) (-14.95 = -14.93 + -0.02)
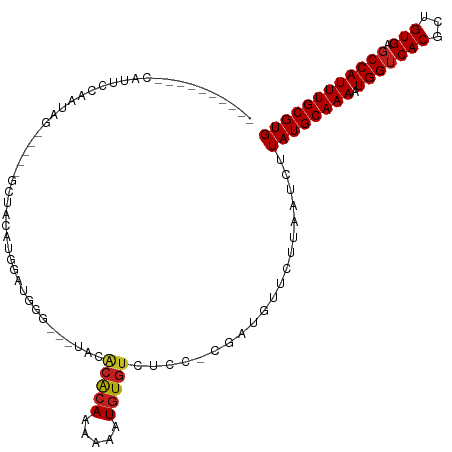
| Location | 7,030,688 – 7,030,784 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 67.85 |
| Shannon entropy | 0.57944 |
| G+C content | 0.50515 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -15.02 |
| Energy contribution | -14.77 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 7030688 96 - 24543557 GUCCAUGUGGCAUGUGGUUCGGUUUUUUGGGGCCUUUGGGAUUGGGAUUAAGUUUGGGUUG-UGGGGAGUCUACCGACAUUCCAAUAGGCUACAUGG ..(((((((((...(((.(((((......((((((..(.((((.((((...)))).)))).-)..)).)))))))))....)))....))))))))) ( -33.50, z-score = -2.29, R) >droSim1.chr3L 6536240 96 - 22553184 GUCCAUGUGGCAUGUGGUUCGGUUUUUUGGGGCCUUUGGGAUU-GGAUUAAGUUUGGUUUGAGGGGGGGUCCACCGACAUUCCAAUAGGCUACAUGG ..(((((((((...(((.(((((......((((((((....(.-.(((((....)))))..)..)))))))))))))....)))....))))))))) ( -35.90, z-score = -2.54, R) >droYak2.chr3L 7616313 94 - 24197627 GUCCAUGUGGCAUGUGGUUCGG-UUUUGGGGGCCUUUGGGAUU-GGAUUAAGUUUUGGGUAUUGGGG-GUCUACCGACAUUCCAAUAGGCUGCAUGG ..(((((..((...(((.((((-(.....(((((((..(..(.-.((......))..)...)..)))-)))))))))....)))....))..))))) ( -36.00, z-score = -3.12, R) >droEre2.scaffold_4784 21433602 92 + 25762168 GUCCGUGUGGCAUGUGGUUCGG-UUUUUGGGACCUUUGGGAUU-GGAUUAAGUUUUGGGUCUGGGGG---CUACCGACAUUCCAAUAGGCUACAUGG ..(((((((((...(((.((((-(((((.((((((..((((((-......)))))))))))).))))---..)))))....)))....))))))))) ( -34.90, z-score = -2.92, R) >droGri2.scaffold_15110 19166234 81 + 24565398 ---------GCCCAGACCGAGGAGGCUUGUGG--UUUGAGUUU-GAGUCAGAUCCGGUUUGGUAGGCUG-CUUUUUGUAUAUUGUUGGUGUUGG--- ---------..(((((((((.((.((..(..(--((((...(.-.(..(......)..)..))))))..-).....))...)).))))).))))--- ( -17.90, z-score = 0.57, R) >consensus GUCCAUGUGGCAUGUGGUUCGG_UUUUUGGGGCCUUUGGGAUU_GGAUUAAGUUUGGGUUGUUGGGG_GUCUACCGACAUUCCAAUAGGCUACAUGG ..(((((((((.((((((((.(.....).)))))..((((((..((...........................))...))))))))).))))))))) (-15.02 = -14.77 + -0.25)
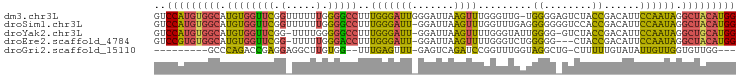
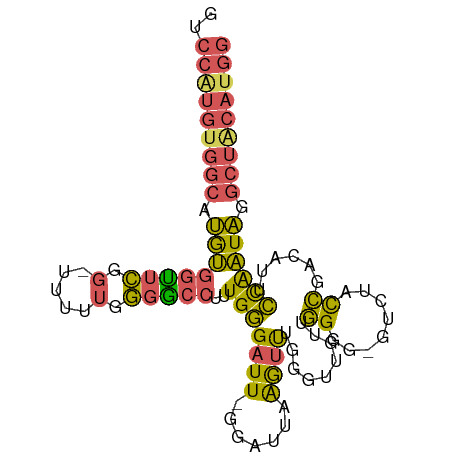
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:43 2011