| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,978,416 – 6,978,486 |
| Length | 70 |
| Max. P | 0.948137 |

| Location | 6,978,416 – 6,978,486 |
|---|---|
| Length | 70 |
| Sequences | 8 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 65.89 |
| Shannon entropy | 0.64129 |
| G+C content | 0.43263 |
| Mean single sequence MFE | -16.05 |
| Consensus MFE | -7.96 |
| Energy contribution | -8.13 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
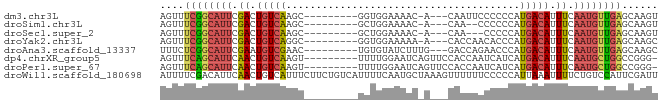
>dm3.chr3L 6978416 70 + 24543557 AGUUUCGGCAUUCGACUGUCAAGC---------GGUGGAAAAC-A---CAAUUCCCCCCAUGACAUUUCAAUGUUGAGCAAGU .(((.(((((((.((.(((((.(.---------((.((((...-.---...)))).))).))))).)).)))))))))).... ( -21.00, z-score = -2.53, R) >droSim1.chr3L 6453396 68 + 22553184 AGUUUCGGCAUUCGACUGUCAAGC---------GCUGGAAAAC-A---CAA--CCCCCCAUGACAUUUCAAUGUUGAGCAAGU .(((.(((((((.((.(((((.(.---------(..((.....-.---...--))..)).))))).)).)))))))))).... ( -14.00, z-score = -0.31, R) >droSec1.super_2 6905384 67 + 7591821 AGUUUCGGCAUUCGACUGUCAAGC---------GCUGGAAAAC-A---CAA---CCCCCAUGACAUUUCAAUGUUGAGCAAGU .(((.(((((((.((.(((((...---------..(((.....-.---...---...)))))))).)).)))))))))).... ( -14.00, z-score = -0.32, R) >droYak2.chr3L 7562049 70 + 24197627 AGUUUCGGCAUUCGACUGUCAGGC---------GGUGGAAAAA-A---CACCAACACCCAUGACAUUUCAAUGUUGAGCAAGC .(((.(((((((.((.(((((((.---------((((......-.---)))).....)).))))).)).)))))))))).... ( -20.50, z-score = -2.04, R) >droAna3.scaffold_13337 3363656 71 - 23293914 UUUCUCGGCAUUCGAAUGUCGAAC---------UGUGUAUCUUUG---GACCAGAACCCAUGACAUUUCAAUGUUGAGCAAGC ...(((((((((.((((((((..(---------((.((.......---.)))))......)))))))).)))))))))..... ( -19.10, z-score = -1.73, R) >dp4.chrXR_group5 680548 73 + 740492 AGUUUCAGCAUUCAACUGUCAAGU---------UUUUGGAAUCAGUUCCACCAAUCAUCAUGACAUUUCAAUGCUGGCCGGG- ....((((((((.((.(((((.((---------(..(((((....)))))..))).....))))).)).)))))))).....- ( -17.80, z-score = -1.92, R) >droPer1.super_67 271628 73 - 321716 AGUUUCAGCAUUCAACUGUCAAGU---------UUUUGGAAUCAGUUCCACCAAUCAUCAUGACAUUUCAAUGCUGGCCGGG- ....((((((((.((.(((((.((---------(..(((((....)))))..))).....))))).)).)))))))).....- ( -17.80, z-score = -1.92, R) >droWil1.scaffold_180698 9067492 83 - 11422946 AUUUUCGACAUUCAACUGUCAUUUCUUCUGUCAUUUUCAAUGCUAAAGUUUUUUCCCCCAUUAAAUUUUCUGUCCAUUCGAUU ....((((((......))))....(((..(.(((.....))))..)))...............................)).. ( -4.20, z-score = -0.02, R) >consensus AGUUUCGGCAUUCGACUGUCAAGC_________GGUGGAAAAC_A___CACCAACCCCCAUGACAUUUCAAUGUUGAGCAAGU ....((((((((.((.(((((.......................................))))).)).))))))))...... ( -7.96 = -8.13 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:33 2011