| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,952,713 – 6,952,804 |
| Length | 91 |
| Max. P | 0.853033 |

| Location | 6,952,713 – 6,952,804 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 70.06 |
| Shannon entropy | 0.56366 |
| G+C content | 0.46988 |
| Mean single sequence MFE | -16.41 |
| Consensus MFE | -7.14 |
| Energy contribution | -7.89 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.853033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6952713 91 - 24543557 UUCAUAUUCCUAUUCCUAUUCCCCUGUCGAUCCGCCACUCGAGGAUCGGUAGUUGCAAUAU-UAAGUCCCAGCAGGACAAUAAUCCCCUCCU .......................((..((((((.........))))))..)).........-...((((.....)))).............. ( -17.80, z-score = -1.62, R) >droSim1.chr3L 6427330 83 - 22553184 ------UUCAUAUUCCUAUUCCCCUGUCGAUCCGCCACUCGAGGAUCGGUAGUUGCAAUAU-UAAGUCCCAGCAGGACA--AAUCCUCUCCU ------.................((..((((((.........))))))..)).........-...((((.....)))).--........... ( -17.80, z-score = -1.56, R) >droSec1.super_2 6879729 83 - 7591821 ------UUCAUAUUCUUAUUCCCCUGUCGAUCCGCCACUCGAGGAUCGGUAGUUGCAAUAU-UAAGUCCCUGCAGGACG--AAUCCCCUCCU ------.....((((........((..((((((.........))))))..)).........-...((((.....)))))--)))........ ( -18.90, z-score = -1.36, R) >droYak2.chr3L 7535797 82 - 24197627 ------UUCAUAUUCCUAUUCCCCUGUCGAUCCGCCACUCGAGGAUCGGUAGUUGCAAUAU-AAGUCCCCAGCAGGAC---AAUCCCCUCCU ------.................((..((((((.........))))))..)).........-..((((......))))---........... ( -18.00, z-score = -1.77, R) >droEre2.scaffold_4784 21355948 80 + 25762168 ------UUCAUAUUCCUAUUCCCCUGCCGAUCCGCCACUCGAGGAUCGGUAGUUGCAAUAU-AAGUCCAC-GCAGGAC---AA-CAUCUCCU ------.................((((((((((.........)))))))))).........-..((((..-...))))---..-........ ( -22.90, z-score = -3.46, R) >dp4.chrXR_group5 653296 75 - 740492 ------------UUCCUAUUACCACGUCGAUCCCCCACUCGAGGAUCGGCAGUUGCAAUAU-UAAACAGU-ACAGCAU---AUGCACAUUCU ------------...............(((((((......).))))))(((..(((..(((-(....)))-)..))).---.)))....... ( -14.70, z-score = -1.90, R) >droPer1.super_67 241651 75 + 321716 ------------UUCCUAUUACCACGUCGAUCCCCCACUCGAGGAUCGGCAGUUGCAAUAU-UAAACCGU-ACAGCAU---AUGCACAUUCU ------------....((((.(.(((((((((((......).)))))))).)).).)))).-........-...((..---..))....... ( -14.20, z-score = -1.60, R) >droMoj3.scaffold_6680 5572191 74 - 24764193 ------UUCCCACAUCUAUUCCUACAACCAGAUGUCGUUC---------CAGUUGCAAUAUCUAAAUACAUAUAUAAC---GAUUGCCGUUG ------.......................((((((.((..---------.....)).))))))...........((((---(.....))))) ( -7.00, z-score = -0.55, R) >consensus ______UUCAUAUUCCUAUUCCCCUGUCGAUCCGCCACUCGAGGAUCGGUAGUUGCAAUAU_UAAACCCC_GCAGGAC___AAUCCCCUCCU .........................((((((((.........)))))))).......................................... ( -7.14 = -7.89 + 0.75)

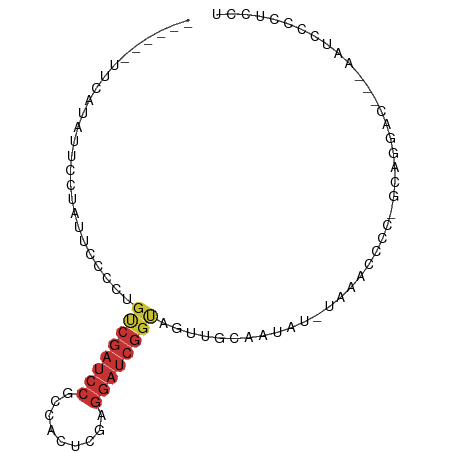

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:31 2011