
| Sequence ID | dm3.chr3L |
|---|---|
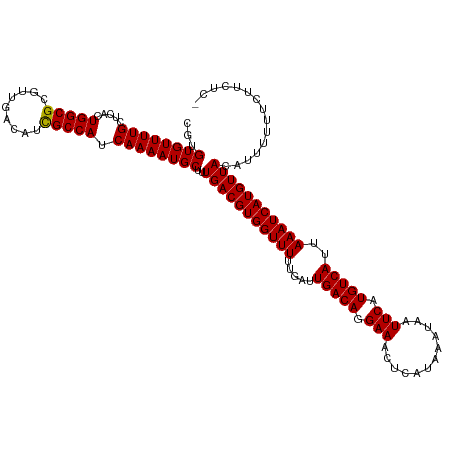
| Location | 6,932,169 – 6,932,304 |
| Length | 135 |
| Max. P | 0.803755 |

| Location | 6,932,169 – 6,932,288 |
|---|---|
| Length | 119 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.91 |
| Shannon entropy | 0.09062 |
| G+C content | 0.36470 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -25.38 |
| Energy contribution | -25.24 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3L 6932169 119 + 24543557 CGUGUGUUUUGCUCACUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUC- ...((((((((.....(((((.........))))).))))))))..(((((((((((.....(((((.(((............))).)))))..)))))))))))..............- ( -25.40, z-score = -0.85, R) >droSim1.chr3L 6405256 119 + 22553184 CGUGUGUUUUGCUCACUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUC- ...((((((((.....(((((.........))))).))))))))..(((((((((((.....(((((.(((............))).)))))..)))))))))))..............- ( -25.40, z-score = -0.85, R) >droSec1.super_2 6858899 119 + 7591821 CGUGUGUUUUGCUCACUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUC- ...((((((((.....(((((.........))))).))))))))..(((((((((((.....(((((.(((............))).)))))..)))))))))))..............- ( -25.40, z-score = -0.85, R) >droYak2.chr3L 7514993 119 + 24197627 CGUGUGUUUUGCUCACUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUC- ...((((((((.....(((((.........))))).))))))))..(((((((((((.....(((((.(((............))).)))))..)))))))))))..............- ( -25.40, z-score = -0.85, R) >droEre2.scaffold_4784 21327995 119 - 25762168 CGUGUGUUUUGCUCACUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACGUUUUUUCUUCUC- (((((((((((.....(((((.........))))).))))))))...((((((((((.....(((((.(((............))).)))))..)))))))))))))............- ( -25.60, z-score = -0.71, R) >droAna3.scaffold_13337 3320874 119 - 23293914 CGUGUGUUUUGCUCAGUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUC- ...((((((((....((((((.........))))))))))))))..(((((((((((.....(((((.(((............))).)))))..)))))))))))..............- ( -27.40, z-score = -1.20, R) >dp4.chrXR_group8 4229005 120 - 9212921 CGUGUGUUUUGCUCAGUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUCUCUUC ...((((((((....((((((.........))))))))))))))..(((((((((((.....(((((.(((............))).)))))..)))))))))))............... ( -27.40, z-score = -1.17, R) >droPer1.super_38 706035 120 + 801819 CGUGUGUUUUGCUCAGUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUCUCUUC ...((((((((....((((((.........))))))))))))))..(((((((((((.....(((((.(((............))).)))))..)))))))))))............... ( -27.40, z-score = -1.17, R) >droWil1.scaffold_180916 2546451 120 - 2700594 CGUGUGUUUUGCUCACUGGCGCGUUGACAUUGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUUUUAUU ...((((((((.....(((((.........))))).))))))))..(((((((((((.....(((((.(((............))).)))))..)))))))))))............... ( -24.10, z-score = -0.32, R) >droVir3.scaffold_13049 7291784 117 + 25233164 CGUGUGUUUUGCUCGCUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUUUC--- ...((((((((.....(((((.........))))).))))))))..(((((((((((.....(((((.(((............))).)))))..)))))))))))............--- ( -25.40, z-score = -0.46, R) >droMoj3.scaffold_6680 5552115 118 + 24764193 CGUGUGUUUUGCUCGCUGGCGCGUUGACAUUGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUCUCU-- ...((((((((.....(((((.........))))).))))))))..(((((((((((.....(((((.(((............))).)))))..))))))))))).............-- ( -24.10, z-score = -0.10, R) >droGri2.scaffold_15110 18982858 114 - 24565398 CAUGUGUUUUGUGCAGUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUU------ ...((((((((....((((((.........))))))))))))))..(((((((((((.....(((((.(((............))).)))))..))))))))))).........------ ( -27.40, z-score = -0.90, R) >consensus CGUGUGUUUUGCUCACUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUC_ ...((((((((.....(((((.........))))).))))))))..(((((((((((.....(((((.(((............))).)))))..)))))))))))............... (-25.38 = -25.24 + -0.14)


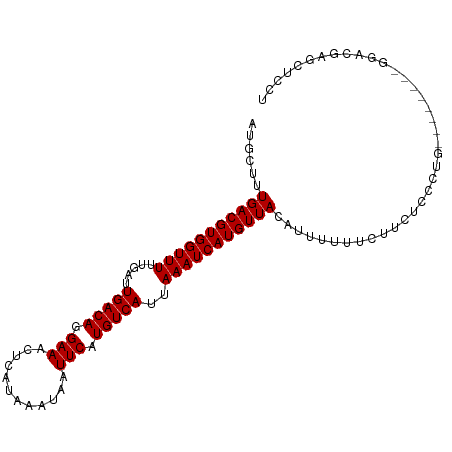
| Location | 6,932,209 – 6,932,304 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 84.59 |
| Shannon entropy | 0.32326 |
| G+C content | 0.33459 |
| Mean single sequence MFE | -18.13 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6932209 95 + 24543557 AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUCUCUG--------GGACGAGCUCCU ..(((((((((((((((.....(((((.(((............))).)))))..)))))))))))..........(((.....--------))).)))).... ( -19.30, z-score = -1.21, R) >droSim1.chr3L 6405296 95 + 22553184 AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUCUCUG--------GGACGAGCUCCU ..(((((((((((((((.....(((((.(((............))).)))))..)))))))))))..........(((.....--------))).)))).... ( -19.30, z-score = -1.21, R) >droSec1.super_2 6858939 95 + 7591821 AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUCUCUG--------GGACGAGCUCCU ..(((((((((((((((.....(((((.(((............))).)))))..)))))))))))..........(((.....--------))).)))).... ( -19.30, z-score = -1.21, R) >droYak2.chr3L 7515033 95 + 24197627 AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUCUCUG--------GGACGAGCUCCU ..(((((((((((((((.....(((((.(((............))).)))))..)))))))))))..........(((.....--------))).)))).... ( -19.30, z-score = -1.21, R) >droEre2.scaffold_4784 21328035 95 - 25762168 AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACGUUUUUUCUUCUCUCUG--------GGACGAGCUCCU ..(((.(((((((((((.....(((((.(((............))).)))))..)))))))))))((((((...........)--------)))))))).... ( -20.00, z-score = -1.31, R) >droAna3.scaffold_13337 3320914 103 - 23293914 AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUCCCAGCUCAGCUCGACCGAGGUCCU ......(((((((((((.....(((((.(((............))).)))))..)))))))))))..........................(((....))).. ( -18.20, z-score = -0.06, R) >dp4.chrXR_group8 4229045 103 - 9212921 AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUCUCUUCUCCUCUCAUCUGGGGCUCCUCCU ......(((((((((((.....(((((.(((............))).)))))..)))))))))))................((((......))))........ ( -19.20, z-score = -1.33, R) >droPer1.super_38 706075 103 + 801819 AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUCUCUUCUCCUCUCAUCUGGGGCUCCUCCU ......(((((((((((.....(((((.(((............))).)))))..)))))))))))................((((......))))........ ( -19.20, z-score = -1.33, R) >droVir3.scaffold_13049 7291824 82 + 25233164 AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUUUCUCCUU--------------------- ......(((((((((((.....(((((.(((............))).)))))..))))))))))).................--------------------- ( -15.20, z-score = -1.77, R) >droMoj3.scaffold_6680 5552155 83 + 24764193 AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUCUCUUCUUU-------------------- ......(((((((((((.....(((((.(((............))).)))))..)))))))))))..................-------------------- ( -15.20, z-score = -1.89, R) >droGri2.scaffold_15110 18982898 79 - 24565398 AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUUAUGC------------------------ ......(((((((((((.....(((((.(((............))).)))))..)))))))))))..............------------------------ ( -15.20, z-score = -1.52, R) >consensus AUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUCUUCUCCCUG________GGACGAGCUCCU ......(((((((((((.....(((((.(((............))).)))))..)))))))))))...................................... (-15.20 = -15.20 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:27 2011