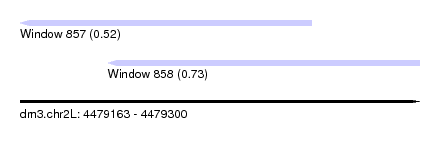
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,479,163 – 4,479,300 |
| Length | 137 |
| Max. P | 0.725769 |

| Location | 4,479,163 – 4,479,263 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Shannon entropy | 0.21686 |
| G+C content | 0.28905 |
| Mean single sequence MFE | -18.91 |
| Consensus MFE | -12.50 |
| Energy contribution | -12.32 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.517756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
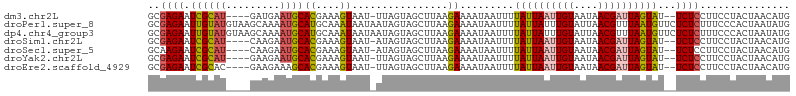
>dm3.chr2L 4479163 100 - 23011544 GCGAGAAUCGCAU----GAUGAAUGCACGAAAGUAAU-UUAGUAGCUUAAGAAAAUAAUUUUAUUAAUUGUAAUAACGAUUAGUAU--UCUCCUUCCUACUAACAUG (((.....)))..----.(((.....((....))...-(((((((....((((........((((((((((....)))))))))))--))).....)))))))))). ( -19.70, z-score = -2.27, R) >droPer1.super_8 1364929 107 + 3966273 GCGAGAAUUGUAUGUAAGCAAAAUGCAUGCAAAUAAUAAUAGUAGCUUAAGAAAAUAAUUUUAUUAUUUGUAUUAACGUUUAAUGUUCUCUCUUUCCCACUAAUAUG ..((((((...((((..((.....))(((((((((((((...((...........))...)))))))))))))..)))).....))))))................. ( -19.00, z-score = -1.64, R) >dp4.chr4_group3 323307 107 + 11692001 GCGAGAAUUGUAUGUAAGCAAAAUGCAUGCAAAUAAUAAUAGUAGCUUAAGAAAAUAAUUUUAUUAUUUGUAUUAACGUUUAAUGUUCUCUCUUUCCCACUAAUAUG ..((((((...((((..((.....))(((((((((((((...((...........))...)))))))))))))..)))).....))))))................. ( -19.00, z-score = -1.64, R) >droSim1.chr2L 4406353 100 - 22036055 GCGAGAAUCGCAU----CAAGAAUGCACGAAAGUAAU-AUAGUAGCUUAAGAAAAUAAUUUUAUUAAUUGUAAUAACGAUUAGUAU--UCUCCUUCCUACUAACAUG (((.....)))..----.........((....))...-.((((((....((((........((((((((((....)))))))))))--))).....))))))..... ( -18.40, z-score = -2.15, R) >droSec1.super_5 2578026 100 - 5866729 GCAAGAAUCGCAU----CAAGAAUGCACGAAAGUAAU-AUAGUAGCUUAAGAAAAUAAUUUUAUUAAUUGUAAUAACGAUUAGUAU--UCUCCUUCCUACUAACAUG .........((((----.....))))((....))...-.((((((....((((........((((((((((....)))))))))))--))).....))))))..... ( -17.60, z-score = -2.17, R) >droYak2.chr2L 4515335 100 - 22324452 GCGAGAAUCGCAU----GAAGAAUGCACGAAAGUAAU-UUAGUAGCUUAAGAAAAUAAUUUUAUUAAUUGUAAUAACGAUUAGUAU--UCUCCUUCCUACUAACAUG (((.....)))((----(........((....))...-(((((((....((((........((((((((((....)))))))))))--))).....)))))))))). ( -19.60, z-score = -2.31, R) >droEre2.scaffold_4929 4569009 100 - 26641161 GCGAGAAUCGCAC----GAAGAAAGCACGAAAGUAAU-UUAGUAGCUUAAGAAAAUAAUUUUAUUAAUUGUAAUAACGAUUAGUAU--UCUCCUUCCUACUAACAUG (((.....)))..----((((.((((((.(((....)-)).)).)))).............((((((((((....)))))))))).--....))))........... ( -19.10, z-score = -2.18, R) >consensus GCGAGAAUCGCAU____GAAGAAUGCACGAAAGUAAU_AUAGUAGCUUAAGAAAAUAAUUUUAUUAAUUGUAAUAACGAUUAGUAU__UCUCCUUCCUACUAACAUG (((.....)))...............((....)).....((((((.....(((........((((((((((....))))))))))........)))))))))..... (-12.50 = -12.32 + -0.18)
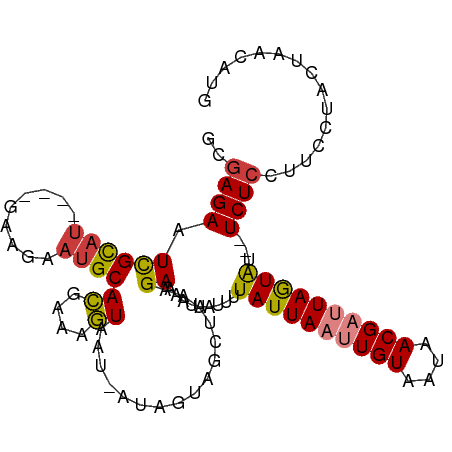
| Location | 4,479,193 – 4,479,300 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.28 |
| Shannon entropy | 0.06009 |
| G+C content | 0.26341 |
| Mean single sequence MFE | -17.77 |
| Consensus MFE | -16.08 |
| Energy contribution | -16.08 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725769 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 4479193 107 - 23011544 GAAAGAUUGAAUUAUUGUAAAAAGCAAAAAA-GACGAAGCGAGAAUCGCAUGAUGAAUGCACGAAAGUAAUUUAGUAGCUUAAGAAAAUAAUUUUAUUAAUUGUAAUA ....(((.((((((((.....((((......-......(((.....)))....(((((..((....)).)))))...)))).....)))))))).))).......... ( -18.40, z-score = -1.94, R) >droSim1.chr2L 4406383 108 - 22036055 GAAAGAUUGAAUUAUUGUAAAAAGCAAAAAAUGACGAAGCGAGAAUCGCAUCAAGAAUGCACGAAAGUAAUAUAGUAGCUUAAGAAAAUAAUUUUAUUAAUUGUAAUA ....(((.((((((((.....((((..........((.(((.....))).))........((....)).........)))).....)))))))).))).......... ( -17.50, z-score = -1.61, R) >droYak2.chr2L 4515365 107 - 22324452 GAAAGAUUGAAUUAUUGUAAAAAGCAAAAAA-GACGAAGCGAGAAUCGCAUGAAGAAUGCACGAAAGUAAUUUAGUAGCUUAAGAAAAUAAUUUUAUUAAUUGUAAUA ....(((.((((((((.....((((......-......(((.....))).....((((..((....)).))))....)))).....)))))))).))).......... ( -17.30, z-score = -1.69, R) >droEre2.scaffold_4929 4569039 107 - 26641161 GAAUGAUUGAAUUAUUGUAAAAAGCAAAAAU-GACGAAGCGAGAAUCGCACGAAGAAAGCACGAAAGUAAUUUAGUAGCUUAAGAAAAUAAUUUUAUUAAUUGUAAUA ...((((.((((((((.....((((......-......(((.....)))((....(((..((....))..))).)).)))).....)))))))).))))......... ( -17.90, z-score = -1.60, R) >consensus GAAAGAUUGAAUUAUUGUAAAAAGCAAAAAA_GACGAAGCGAGAAUCGCAUGAAGAAUGCACGAAAGUAAUUUAGUAGCUUAAGAAAAUAAUUUUAUUAAUUGUAAUA ....(((.((((((((.....((((.............(((.....)))...........((....)).........)))).....)))))))).))).......... (-16.08 = -16.08 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:36 2011