| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,919,172 – 6,919,268 |
| Length | 96 |
| Max. P | 0.768769 |

| Location | 6,919,172 – 6,919,268 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 69.52 |
| Shannon entropy | 0.60173 |
| G+C content | 0.46106 |
| Mean single sequence MFE | -21.99 |
| Consensus MFE | -9.76 |
| Energy contribution | -9.84 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
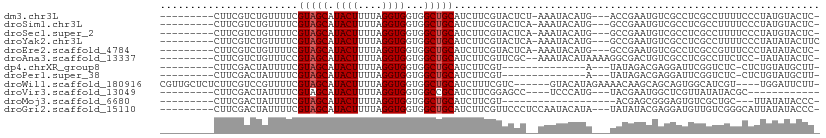
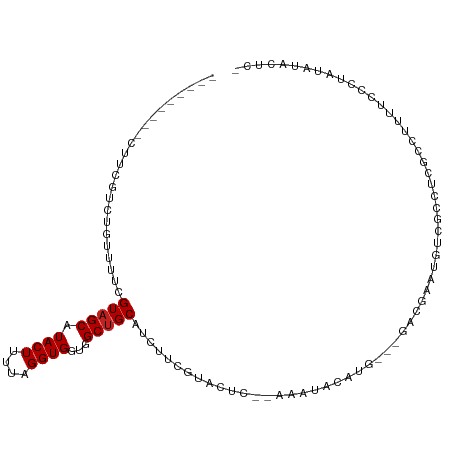
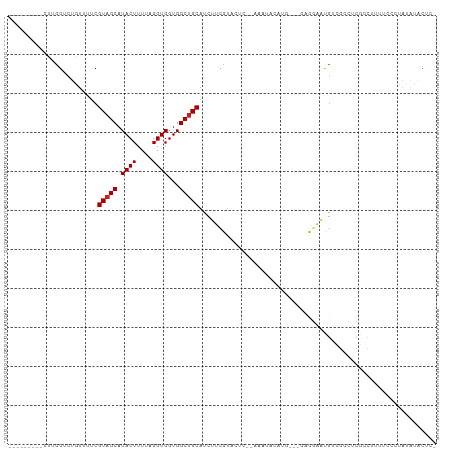
>dm3.chr3L 6919172 96 + 24543557 ---------CUUCGUCUGUUUUCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUCGUACUCU-AAAUACAUG---ACCGAAUGUCGCCUCGCCUUUUCCCUAUGUACUC- ---------..............(((.((((.....((((((.(((.((((..(((..(...-........)---..))))))).)))))))))......)))))))..- ( -20.10, z-score = -1.42, R) >droSim1.chr3L 6392574 96 + 22553184 ---------CUUCGUCUGUUUUCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUCGUACUCA-AAAUACAUG---GCCGAAUGUCGCCUCGCCUUUUCCCUAUGUACUC- ---------..............(((.((((.....((((((.(((.((((..(((..(...-........)---..))))))).)))))))))......)))))))..- ( -21.70, z-score = -1.57, R) >droSec1.super_2 6841134 96 + 7591821 ---------CUUCGUCUGUUUUCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUCGUACUCA-AAAUACAUG---GCCGAAUGUCGCCUCGCCUUUUCCCUAUGUACUC- ---------..............(((.((((.....((((((.(((.((((..(((..(...-........)---..))))))).)))))))))......)))))))..- ( -21.70, z-score = -1.57, R) >droYak2.chr3L 7501078 97 + 24197627 ---------CUUCGUCUGUUUUCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUCGUACUCA-AAAUACAUG---GCCGAAUGUCGCCUCGCCUUUUCCCUAUAUACUUC ---------....((.((((.....)))).))....((((((.(((.((((..(((..(...-........)---..))))))).)))))))))................ ( -19.80, z-score = -1.21, R) >droEre2.scaffold_4784 21314123 96 - 25762168 ---------CUUCGUCUGUUUUCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUCGUACUCA-AAAUACAUG---GCCGAAUGUCGCCUCGCCGUUUCCCUAUAUACUC- ---------....((.((((.....)))).)).....(((((.(((.((((..(((..(...-........)---..))))))).))))))))................- ( -19.80, z-score = -0.71, R) >droAna3.scaffold_13337 3305812 97 - 23293914 ---------CUUCGUCUGUUUCCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUCGUUCGC--AAAUACAUAAAAGGCGACUGUCGCCUCGCCUUCUCC-UAUAUACUC- ---------....((.((((.....)))).))...((((.((.((((((..........))--)..........(((((.....))))).))).)).))-)).......- ( -22.20, z-score = -1.49, R) >dp4.chrXR_group8 4214702 82 - 9212921 ---------CUUCGACUAUUUUCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUCGU--------------A---UAUAGACGAGGAUUCGGUCUC-CUCUGUAUGCUU- ---------...(((......))).(((((((.....((.((.(((((.((((((((--------------.---.....)))))))).))))).)-).))))))))).- ( -28.60, z-score = -3.97, R) >droPer1.super_38 691620 82 + 801819 ---------CUUCGACUAUUUUCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUCGU--------------A---UAUAGACGAGGAUUCGGUCUC-CUCUGUAUGCUU- ---------...(((......))).(((((((.....((.((.(((((.((((((((--------------.---.....)))))))).))))).)-).))))))))).- ( -28.60, z-score = -3.97, R) >droWil1.scaffold_180916 2508787 99 - 2700594 CGUUGCUCUCUUCGUCCGUUUUCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUUCGUC------GUACAUAGAAAACAAGCAGCAGUGGCAUCGU----UGGAUUCUU- .((..(((((((.....(((((((((((.((((....))))...))))).......(..------...)...))))))))).)).))..)).....----.........- ( -21.30, z-score = 0.31, R) >droVir3.scaffold_13049 7275468 82 + 25233164 ---------CUUCGACUAUUUUCGUAGCAUACUUUUAGGUGGUGGCCGCAUCUUCGGAGCC----UCCCAUG---UACGAAUGGCUCGUUAUAUACGC------------ ---------...(((((((..(((((.((((((....)))((.(((....((....)))))----.)).)))---))))))))).)))..........------------ ( -18.40, z-score = 0.22, R) >droMoj3.scaffold_6680 5533326 78 + 24764193 ---------CUUCGACUAUUUUCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUCGU-------------------ACGAGCGGGAGUGUCGCUGC---UUAUAUACCC- ---------..............(((((.((((....))))...)))))......((-------------------(..((((((.....).))))---)..)))....- ( -18.60, z-score = 0.18, R) >droGri2.scaffold_15110 18968881 97 - 24565398 ---------CUUCGACUAUUUUCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUCGUUCCCUCCAAUACAUA---UAUAUACGAGGAUGUUGUCGGGCAUUAUAUACCC- ---------.((((((.......(((((.((((....))))...)))))((((((((...............---.....))))))))...))))))............- ( -23.05, z-score = -1.07, R) >consensus _________CUUCGUCUGUUUUCGUAGCAUACUUUUAGGUGGUGGCUGCAUCUUCGUACUC__AAAUACAUG___GACGAAUGUCGCCUCGCCUUUUCCCUAUAUACUC_ .......................(((((.((((....))))...)))))............................................................. ( -9.76 = -9.84 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:24 2011