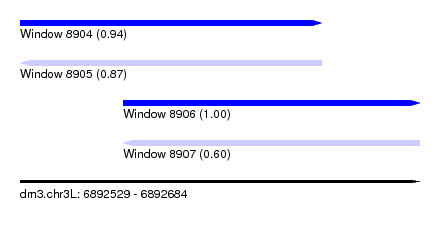
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,892,529 – 6,892,684 |
| Length | 155 |
| Max. P | 0.997115 |

| Location | 6,892,529 – 6,892,646 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.12 |
| Shannon entropy | 0.19206 |
| G+C content | 0.45253 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -19.32 |
| Energy contribution | -22.14 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
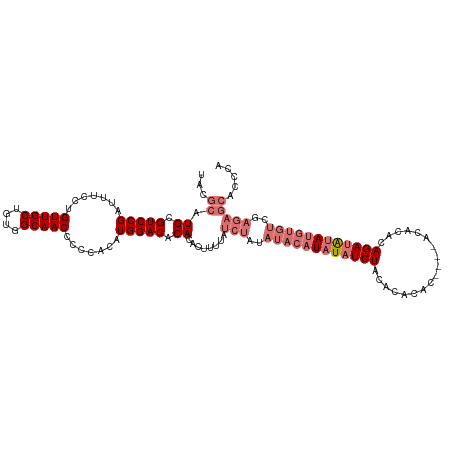
>dm3.chr3L 6892529 117 + 24543557 UACGCAUGCGUCCGAUUUCCUGUUGCUGUGGCAACCCCCACAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACACACAAACACACAGAUAUAUGUGUCGAGAGCACCCA ...((.((.(((((.......(((((....))))).......))))).))........(((..((((((((((((..................))))))))))))..)))))..... ( -29.01, z-score = -3.21, R) >droSim1.chr3L 6365579 113 + 22553184 UACGCAUGCGUCCGAUUUCCUGUUGCUGUGGCAACCCCCACAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACAC----ACACACAGAUAUAUGUGUCGAGAGCACCCA ...((.((.(((((.......(((((....))))).......))))).))........(((..((((((((((((........----......))))))))))))..)))))..... ( -29.28, z-score = -3.23, R) >droSec1.super_2 6814647 97 + 7591821 UACGCAUGCGUCCGAUUUCCUGUUGCUGUGGCAACCCCCACAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACAC----ACACACAGA-GCACC--------------- ...((.((.(((((.......(((((....))))).......))))).))..........((((....))))...........----.........-))...--------------- ( -14.64, z-score = -0.55, R) >droEre2.scaffold_4784 21286637 111 - 25762168 UACGCAUGCGUCCGAUUUCCUGUUGCUGUGGCAACCCCCACAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACAC------ACACACAGAUAUAUGUGUCGAGAGCAGGCA ...((.((((((((.......(((((....))))).......)))))...........(((..((((((((((((......------......))))))))))))..)))))).)). ( -33.04, z-score = -3.54, R) >consensus UACGCAUGCGUCCGAUUUCCUGUUGCUGUGGCAACCCCCACAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACAC____ACACACAGAUAUAUGUGUCGAGAGCACCCA ...((.((.(((((.......(((((....))))).......))))).))........(((..((((((((((((..................))))))))))))..)))))..... (-19.32 = -22.14 + 2.81)

| Location | 6,892,529 – 6,892,646 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.12 |
| Shannon entropy | 0.19206 |
| G+C content | 0.45253 |
| Mean single sequence MFE | -36.79 |
| Consensus MFE | -31.47 |
| Energy contribution | -32.09 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6892529 117 - 24543557 UGGGUGCUCUCGACACAUAUAUCUGUGUGUUUGUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGCAACAGGAAAUCGGACGCAUGCGUA .(.((((.....((((((.((((((((((..(((((........)))))..))))))))))...))))))((((........(((((....)))))........)))))))).)... ( -36.19, z-score = -0.91, R) >droSim1.chr3L 6365579 113 - 22553184 UGGGUGCUCUCGACACAUAUAUCUGUGUGU----GUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGCAACAGGAAAUCGGACGCAUGCGUA .(.((((.....((((((.((((((((((.----.(((((....)))))..))))))))))...))))))((((........(((((....)))))........)))))))).)... ( -39.39, z-score = -2.12, R) >droSec1.super_2 6814647 97 - 7591821 ---------------GGUGC-UCUGUGUGU----GUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGCAACAGGAAAUCGGACGCAUGCGUA ---------------..(((-((((((((.----.(((((....)))))..)))))))).....((((((((((........(((((....)))))........))))))))))))) ( -34.09, z-score = -2.59, R) >droEre2.scaffold_4784 21286637 111 + 25762168 UGCCUGCUCUCGACACAUAUAUCUGUGUGU------GUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGCAACAGGAAAUCGGACGCAUGCGUA .((.(((.....((((((.((((((((((.------.((((....))))..))))))))))...))))))((((........(((((....)))))........))))))).))... ( -37.49, z-score = -1.62, R) >consensus UGGGUGCUCUCGACACAUAUAUCUGUGUGU____GUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGCAACAGGAAAUCGGACGCAUGCGUA ..............(((((((((((..((........))..)))))))))))............((((((((((........(((((....)))))........))))))))))... (-31.47 = -32.09 + 0.62)

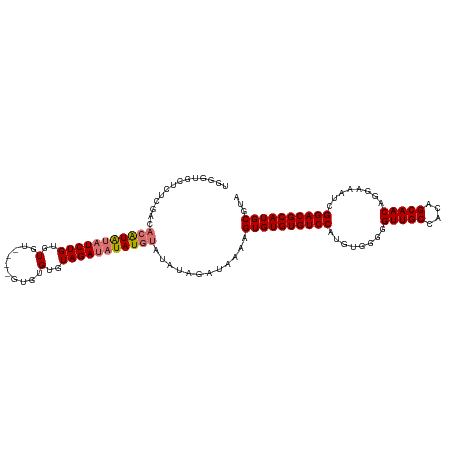
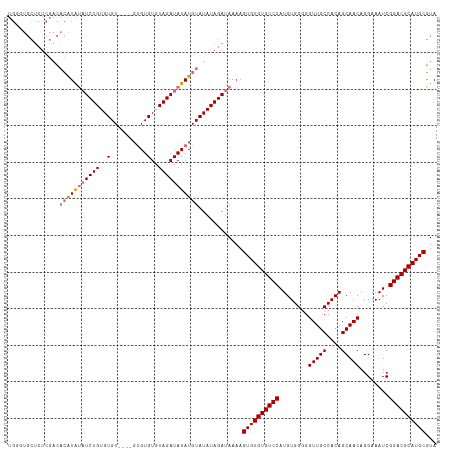
| Location | 6,892,569 – 6,892,684 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 95.31 |
| Shannon entropy | 0.06388 |
| G+C content | 0.34927 |
| Mean single sequence MFE | -19.00 |
| Consensus MFE | -17.87 |
| Energy contribution | -17.87 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.997115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6892569 115 + 24543557 CAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACACACAAACACACAGAUAUAUGUGUCGAGAGCACCCAACGCAAUAUUUGAAUAUUCACGCAUACCAUAAACUAAA .((((.............(((..((((((((((((..................))))))))))))..)))((.......))......................))))........ ( -17.87, z-score = -3.34, R) >droSim1.chr3L 6365619 111 + 22553184 CAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACACAC----ACACAGAUAUAUGUGUCGAGAGCACCCAACGCAAUAUUUGAAUAUUCACGCAUACCAUAAACUAAA .((((.............(((..((((((((((((..........----....))))))))))))..)))((.......))......................))))........ ( -18.14, z-score = -3.45, R) >droEre2.scaffold_4784 21286677 109 - 25762168 CAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACACAC------ACAGAUAUAUGUGUCGAGAGCAGGCAACGCAAUAUUUGAAUAUUCACGCAUACCAUAAACUAAA .((((.............(((..((((((((((((..........------..))))))))))))..)))((.(....)(.(((((....))))))..))...))))........ ( -21.00, z-score = -3.72, R) >consensus CAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACACAC____ACACAGAUAUAUGUGUCGAGAGCACCCAACGCAAUAUUUGAAUAUUCACGCAUACCAUAAACUAAA .((((.............(((..((((((((((((..................))))))))))))..)))((.......))......................))))........ (-17.87 = -17.87 + -0.00)

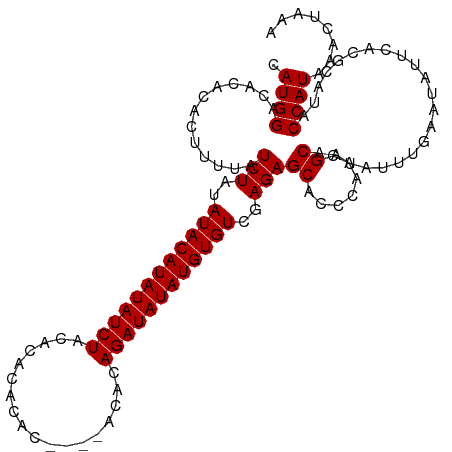
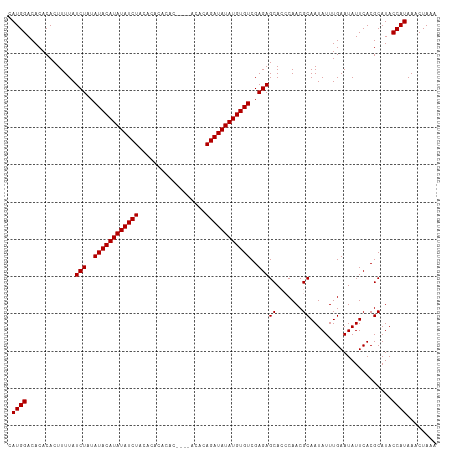
| Location | 6,892,569 – 6,892,684 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 95.31 |
| Shannon entropy | 0.06388 |
| G+C content | 0.34927 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -23.67 |
| Energy contribution | -24.67 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6892569 115 - 24543557 UUUAGUUUAUGGUAUGCGUGAAUAUUCAAAUAUUGCGUUGGGUGCUCUCGACACAUAUAUCUGUGUGUUUGUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUG .......(((((....((.((.(((((((........))))))).)).))((((((.((((((((((..(((((........)))))..))))))))))...))))))..))))) ( -28.10, z-score = -1.15, R) >droSim1.chr3L 6365619 111 - 22553184 UUUAGUUUAUGGUAUGCGUGAAUAUUCAAAUAUUGCGUUGGGUGCUCUCGACACAUAUAUCUGUGU----GUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUG .......(((((....((.((.(((((((........))))))).)).))((((((.(((((((((----(..(((((....)))))..))))))))))...))))))..))))) ( -31.30, z-score = -2.43, R) >droEre2.scaffold_4784 21286677 109 + 25762168 UUUAGUUUAUGGUAUGCGUGAAUAUUCAAAUAUUGCGUUGCCUGCUCUCGACACAUAUAUCUGU------GUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUG ...(((....((((.((((.(((((....))))))))))))).)))...(((((((((((((((------(((..((((....))))..)))))))))....))))))))).... ( -32.00, z-score = -3.12, R) >consensus UUUAGUUUAUGGUAUGCGUGAAUAUUCAAAUAUUGCGUUGGGUGCUCUCGACACAUAUAUCUGUGU____GUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUG .......((((((((((((.(((((....)))))))))...))))....(((((((((((((((((....((((((......))))))..))))))))....))))))))))))) (-23.67 = -24.67 + 1.00)

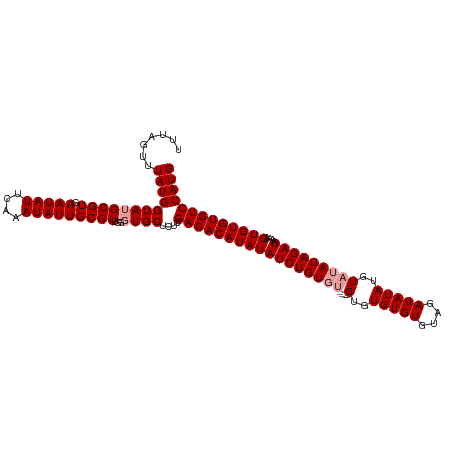
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:22 2011