
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,891,092 – 6,891,185 |
| Length | 93 |
| Max. P | 0.998346 |

| Location | 6,891,092 – 6,891,185 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 85.20 |
| Shannon entropy | 0.29299 |
| G+C content | 0.47601 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -22.07 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.996188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6891092 93 + 24543557 GCGGCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGGUUCGCUUUUCCUGCGCUUU----------- ..(((.((((.((((((((....((.((((....((((.(((((((...))))))).))))..)))).))....))))))))..)))))))..----------- ( -34.30, z-score = -3.48, R) >droSim1.chr3L 6364156 93 + 22553184 GCGGCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGGUUCGCUUUUCCUGCGCUUU----------- ..(((.((((.((((((((....((.((((....((((.(((((((...))))))).))))..)))).))....))))))))..)))))))..----------- ( -34.30, z-score = -3.48, R) >droSec1.super_2 6813230 93 + 7591821 GCGGCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGGUUCGCUUUUCCUGCGCUUU----------- ..(((.((((.((((((((....((.((((....((((.(((((((...))))))).))))..)))).))....))))))))..)))))))..----------- ( -34.30, z-score = -3.48, R) >droYak2.chr3L 7469623 93 + 24197627 GCGGCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGGUUCGCUUUUCCUGCGCUUU----------- ..(((.((((.((((((((....((.((((....((((.(((((((...))))))).))))..)))).))....))))))))..)))))))..----------- ( -34.30, z-score = -3.48, R) >droEre2.scaffold_4784 21285257 93 - 25762168 GCGGCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGGUUCGCUUUUCCUGCGCUUU----------- ..(((.((((.((((((((....((.((((....((((.(((((((...))))))).))))..)))).))....))))))))..)))))))..----------- ( -34.30, z-score = -3.48, R) >droAna3.scaffold_13337 3267290 86 - 23293914 GCGCCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGGUUCGCUUCGCCC------------------ (((..((((((..((.(((((((((((...))))((((.(((((((...))))))).))))))))))).))..))).))).)))..------------------ ( -30.70, z-score = -3.34, R) >droPer1.super_38 661491 87 + 801819 GCGGCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGCUUCGCCGUGCCAC----------------- ..((((.(.((.((((((((((.((((...))))((((.(((((((...))))))).))))..)))))...))))).)).)))))..----------------- ( -33.30, z-score = -3.49, R) >dp4.chrXR_group8 4184814 87 - 9212921 GCGGCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGCUUCGCCGUGCCAC----------------- ..((((.(.((.((((((((((.((((...))))((((.(((((((...))))))).))))..)))))...))))).)).)))))..----------------- ( -33.30, z-score = -3.49, R) >droWil1.scaffold_180916 2450695 104 - 2700594 GCAGCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGACUACAAACGGCCCCAAGCCGCUCGCCCUGC ((((..(((((..((.(((((((((((...))))((((.(((((((...))))))).))))))))))).))...........(((.....)))))).)).)))) ( -34.80, z-score = -2.93, R) >droGri2.scaffold_15110 18937427 87 - 24565398 AGCGCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACAUGGUUACCAAGUUUGCGCAUUUUUCACUCGACUCGCUGCCAGCU----------------- ((((((((((...((.(((((((((((...))))((((.(((.(((...))).))).))))))))))).))...)).)))))..)))----------------- ( -27.60, z-score = -1.90, R) >droMoj3.scaffold_6680 5499727 87 + 24764193 AGCGCAGCAGAAGAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGACUCGCUGCCAGCU----------------- ((((((((((..(((.(((((((((((...))))((((.(((((((...))))))).))))))))))).)))..)).)))))..)))----------------- ( -37.00, z-score = -4.90, R) >droVir3.scaffold_13049 7239669 87 + 25233164 AGCGCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGACUCGCUGCCAGCU----------------- ((((((((((...((.(((((((((((...))))((((.(((((((...))))))).))))))))))).))...)).)))))..)))----------------- ( -31.70, z-score = -3.16, R) >consensus GCGGCAGCAGCAAAGCGGAAAAUGUUAAAAUAACGCGCUAACUUGGUUACCAAGUUUGCGCAUUUUUCACUCGGUUCGCUUUUCCCG_________________ ...((.((......))(((((((((((...))))((((.(((((((...))))))).))))))))))).........))......................... (-22.07 = -22.40 + 0.33)

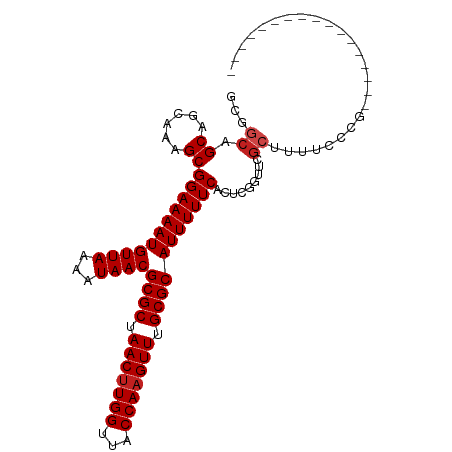
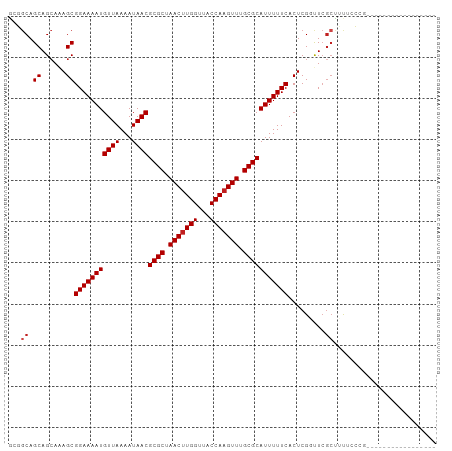
| Location | 6,891,092 – 6,891,185 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 85.20 |
| Shannon entropy | 0.29299 |
| G+C content | 0.47601 |
| Mean single sequence MFE | -36.76 |
| Consensus MFE | -24.01 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.33 |
| SVM RNA-class probability | 0.998346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6891092 93 - 24543557 -----------AAAGCGCAGGAAAAGCGAACCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGCCGC -----------...((((((..((((((.......((((((((((((.(((((((...))))))).))))))).)))))........)))))).)))).))... ( -34.46, z-score = -3.38, R) >droSim1.chr3L 6364156 93 - 22553184 -----------AAAGCGCAGGAAAAGCGAACCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGCCGC -----------...((((((..((((((.......((((((((((((.(((((((...))))))).))))))).)))))........)))))).)))).))... ( -34.46, z-score = -3.38, R) >droSec1.super_2 6813230 93 - 7591821 -----------AAAGCGCAGGAAAAGCGAACCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGCCGC -----------...((((((..((((((.......((((((((((((.(((((((...))))))).))))))).)))))........)))))).)))).))... ( -34.46, z-score = -3.38, R) >droYak2.chr3L 7469623 93 - 24197627 -----------AAAGCGCAGGAAAAGCGAACCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGCCGC -----------...((((((..((((((.......((((((((((((.(((((((...))))))).))))))).)))))........)))))).)))).))... ( -34.46, z-score = -3.38, R) >droEre2.scaffold_4784 21285257 93 + 25762168 -----------AAAGCGCAGGAAAAGCGAACCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGCCGC -----------...((((((..((((((.......((((((((((((.(((((((...))))))).))))))).)))))........)))))).)))).))... ( -34.46, z-score = -3.38, R) >droAna3.scaffold_13337 3267290 86 + 23293914 ------------------GGGCGAAGCGAACCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGGCGC ------------------.(((((((((.......((((((((((((.(((((((...))))))).))))))).)))))........)))))))))........ ( -33.36, z-score = -3.11, R) >droPer1.super_38 661491 87 - 801819 -----------------GUGGCACGGCGAAGCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGCCGC -----------------((((((((((((((((..((((((((((((.(((((((...))))))).))))))).)))))........)))))))))).)))))) ( -43.20, z-score = -5.92, R) >dp4.chrXR_group8 4184814 87 + 9212921 -----------------GUGGCACGGCGAAGCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGCCGC -----------------((((((((((((((((..((((((((((((.(((((((...))))))).))))))).)))))........)))))))))).)))))) ( -43.20, z-score = -5.92, R) >droWil1.scaffold_180916 2450695 104 + 2700594 GCAGGGCGAGCGGCUUGGGGCCGUUUGUAGUCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGCUGC ((((.(((((((((.....)))))))))(((.(((((.(((((((((.(((((((...))))))).))))((((...))))))))).))))).))).))))... ( -44.90, z-score = -3.48, R) >droGri2.scaffold_15110 18937427 87 + 24565398 -----------------AGCUGGCAGCGAGUCGAGUGAAAAAUGCGCAAACUUGGUAACCAUGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGCGCU -----------------(((..(((((.(((.(((((.(((((((((.(((.(((...))).))).))))((((...))))))))).))))).))))))))))) ( -32.00, z-score = -2.35, R) >droMoj3.scaffold_6680 5499727 87 - 24764193 -----------------AGCUGGCAGCGAGUCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUCUUCUGCUGCGCU -----------------(((..(((((.((..(((((.(((((((((.(((((((...))))))).))))((((...))))))))).)))))..)))))))))) ( -36.00, z-score = -4.13, R) >droVir3.scaffold_13049 7239669 87 - 25233164 -----------------AGCUGGCAGCGAGUCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGCGCU -----------------(((..(((((.(((.(((((.(((((((((.(((((((...))))))).))))((((...))))))))).))))).))))))))))) ( -36.10, z-score = -3.66, R) >consensus _________________CAGGAAAAGCGAACCGAGUGAAAAAUGCGCAAACUUGGUAACCAAGUUAGCGCGUUAUUUUAACAUUUUCCGCUUUGCUGCUGCCGC ........................((((....(((((.(((((((((.(((((((...))))))).))))((((...))))))))).)))))...))))..... (-24.01 = -24.22 + 0.22)

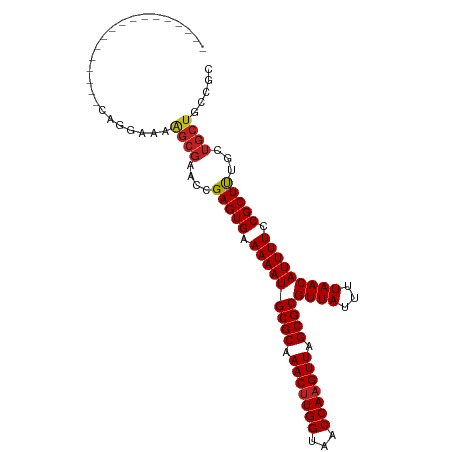
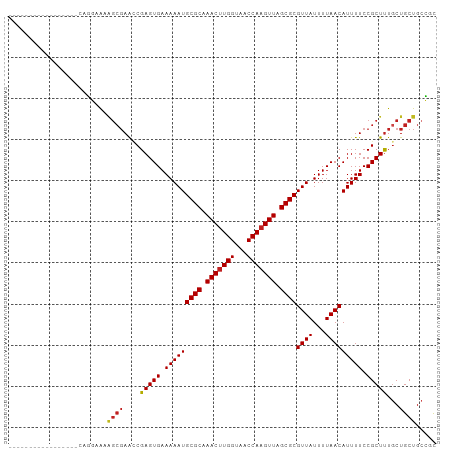
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:19 2011