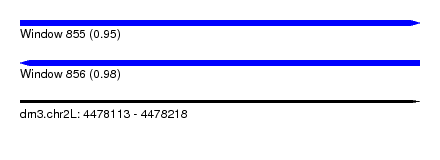
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,478,113 – 4,478,218 |
| Length | 105 |
| Max. P | 0.981164 |

| Location | 4,478,113 – 4,478,218 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.66 |
| Shannon entropy | 0.63971 |
| G+C content | 0.53843 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -12.54 |
| Energy contribution | -11.63 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
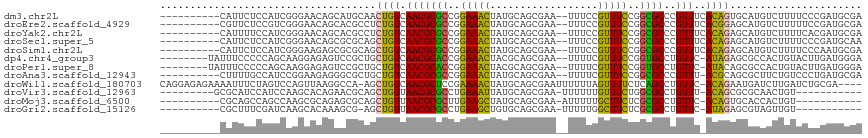
>dm3.chr2L 4478113 105 + 23011544 UCGCAUCGGGAAAAGACAUGCACUGUGAAACGGGCGCCGGAAACGGAAA--UUCGCUGCAUAGUUUCCGGCGCGUUGACAGUUGCAUGCUGUUCCCGAUGGAGAAUG---------- ((.(((((((((.((.((((((((((.((....(((((((((((.....--...........))))))))))).)).)))).)))))))).))))))))))).....---------- ( -53.89, z-score = -6.38, R) >droEre2.scaffold_4929 4567964 105 + 26641161 UCGCAUCGGAAAAAGACAUGCUCCGUGAAACGGGCGCCGGAAACGGAAA--UUCGCUGCAUAGUUUCCGGCGCGUUGACAGAGGCGUGCUGUUCCCGACGGAGAACG---------- ((.(.((((.((.((.(((((((((.....)))(((((((((((.....--...........))))))))))).........)))))))).)).)))).))).....---------- ( -39.59, z-score = -1.27, R) >droYak2.chr2L 4514294 105 + 22324452 UCGCAUCGUGAAAAGACAUGCUCUGUGAAACGGGCGCCGGAAACGGAAA--UUCGCUGCAUAGUUUCCGGCGCGUUGACAGAGGCGUGCUGUUCCCGAUGGAAAAUG---------- ((.(((((.(((.((.((((((((((.((....(((((((((((.....--...........))))))))))).)).))))).))))))).))).))))))).....---------- ( -46.59, z-score = -4.20, R) >droSec1.super_5 2576985 105 + 5866729 UUGCAUCGGGAAAAGACAUGCUCUGUGAAACGGGCGCCGGAAACGGAAA--UUCGCUGCAUAGUUUCCGGCGCGUUGACAGCUGCGCGCUGUUCCCGAUGGAGAAUG---------- ((.(((((((((.((.(.(((.((((.((....(((((((((((.....--...........))))))))))).)).))))..))).))).))))))))).))....---------- ( -46.59, z-score = -3.72, R) >droSim1.chr2L 4405312 105 + 22036055 UCGCAUUGGGAAAAGACAUGCUCUGUGAAACGGGCGCCGGAAACGGAAA--UUCGCUGCAUAGUUUCCGGCGCGUUGACAGCUGCGCGCUCUUCCCGAUGGAGAAUG---------- ((.(((((((((.((.(.(((.((((.((....(((((((((((.....--...........))))))))))).)).))))..))).))).))))))))))).....---------- ( -44.79, z-score = -3.23, R) >dp4.chr4_group3 322230 106 - 11692001 UCCCAUCAAGUACAGUGGCGCUCUAU-GAACAGGCACCGGAAACGAAAA--UUCGCUGCGUAGUUUCCGGUGCGUUGACAGCAGCGGACUCUCCUUGCUGGGGGAAAUA-------- ((((.((((((...((((....))))-..))..(((((((((((.....--...........))))))))))).))))((((((.((.....)))))))).))))....-------- ( -39.09, z-score = -2.03, R) >droPer1.super_8 1363852 106 - 3966273 UCCCAUCAAGUACAGUGGCGCUGUAU-GAACAGGCACCGGAAACGAAAA--UUCGCUGCGUAGUUUCCGGUGCGUUGACAGCAGCGGACUCUCCUUGCUGGGGGAAAUA-------- ((((.((((((((((.....))))))-......(((((((((((.....--...........))))))))))).))))((((((.((.....)))))))).))))....-------- ( -44.39, z-score = -3.41, R) >droAna3.scaffold_12943 1678168 104 - 5039921 UCGCAUCAGGGACAGAAGCGCUGCGU-AAACGGGCGCCGGAAACGAAAA--UUCGCUGCAUAGUUUCCGGCGCGUUGACAGCAGCGCCCUCUUCCGGAUGGCAAAAG---------- ..(((((..(((.(((.(((((((((-.(((..(((((((((((.....--...........)))))))))))))).)).)))))))..)))))).))).)).....---------- ( -47.19, z-score = -3.51, R) >droWil1.scaffold_180703 1222562 111 - 3946847 ----UCGCAGAUCAAGAUCAUUCUGU-GAACAGGCUGAGAAAACUAAAAAAUUCGCUGCAUAGUUUUCGGAGCGUUGACAGCU-UGGCCUUAACUGGACUAGAAAUUUUCUCUCCUG ----(((((((..........)))))-)).((((..(((((((...........((((((..((((...))))..)).))))(-(((((......)).))))...))))))).)))) ( -30.40, z-score = -1.58, R) >droVir3.scaffold_12963 8392800 95 - 20206255 -----------ACAGUUGCGCGCUGU-AAACAGGCGCCAGAAACAAAAAA-UUCGCUGCAUAAUUUCAGGCGCGUUAACAGCUGCGUUCUGUGCUUGGAUGGAUGCGC--------- -----------...(((((((.(((.-...)))))))....)))......-...((.((((...((((((((((..(((......))).))))))))))...))))))--------- ( -29.20, z-score = 0.32, R) >droMoj3.scaffold_6500 11836437 94 - 32352404 -----------ACAGUGGUGCACUGU-GAACAGGCGCGAGAAGCAAAAAU-UUCGCUGCAUAGCUUCAAGCGCGUUAACAGCUGCGCUCUGCGCUUGGCUGGCUGCG---------- -----------.((((.((((.(((.-...)))((((((((........)-))))).))..........)))).....(((((((((...))))..)))))))))..---------- ( -33.90, z-score = 0.77, R) >droGri2.scaffold_15126 3030152 93 - 8399593 -----------ACAACUACGCUCUAU-GAACAGGCGCGAGAAGCCAAAAA-UUCGCUGCACAGCUUCAGGCGCGUUAACAGCU-CGCUUUGUGCUUGAUCGAAAGCG---------- -----------.......((((...(-((.((((((((((((((......-...........))))).((((.((.....)).-))))))))))))).)))..))))---------- ( -28.03, z-score = -0.42, R) >consensus UCGCAUCGGGAACAGACACGCUCUGU_AAACAGGCGCCGGAAACGAAAA__UUCGCUGCAUAGUUUCCGGCGCGUUGACAGCUGCGCGCUGUUCCCGAUGGAGAAUG__________ ..................((((......(((..((((..(((((..................)))))..)))))))......))))............................... (-12.54 = -11.63 + -0.91)

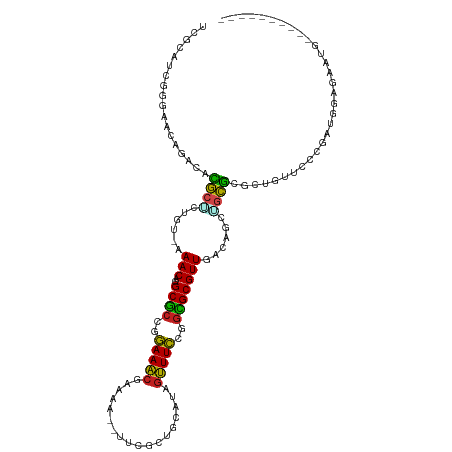
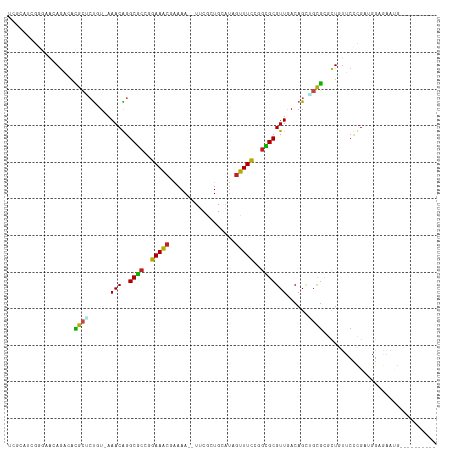
| Location | 4,478,113 – 4,478,218 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.66 |
| Shannon entropy | 0.63971 |
| G+C content | 0.53843 |
| Mean single sequence MFE | -38.73 |
| Consensus MFE | -11.12 |
| Energy contribution | -10.14 |
| Covariance contribution | -0.98 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4478113 105 - 23011544 ----------CAUUCUCCAUCGGGAACAGCAUGCAACUGUCAACGCGCCGGAAACUAUGCAGCGAA--UUUCCGUUUCCGGCGCCCGUUUCACAGUGCAUGUCUUUUCCCGAUGCGA ----------.....(((((((((((.((((((((.((((.((((((((((((((...........--.....)))))))))))..)))..)))))))))).)).))))))))).)) ( -50.89, z-score = -7.14, R) >droEre2.scaffold_4929 4567964 105 - 26641161 ----------CGUUCUCCGUCGGGAACAGCACGCCUCUGUCAACGCGCCGGAAACUAUGCAGCGAA--UUUCCGUUUCCGGCGCCCGUUUCACGGAGCAUGUCUUUUUCCGAUGCGA ----------.....(((((((((((.((((.((.(((((.((((((((((((((...........--.....)))))))))))..)))..))))))).)).)).))))))))).)) ( -42.79, z-score = -3.13, R) >droYak2.chr2L 4514294 105 - 22324452 ----------CAUUUUCCAUCGGGAACAGCACGCCUCUGUCAACGCGCCGGAAACUAUGCAGCGAA--UUUCCGUUUCCGGCGCCCGUUUCACAGAGCAUGUCUUUUCACGAUGCGA ----------.....(((((((((((..(((.((.(((((.((((((((((((((...........--.....)))))))))))..)))..))))))).)))..)))).))))).)) ( -40.69, z-score = -4.12, R) >droSec1.super_5 2576985 105 - 5866729 ----------CAUUCUCCAUCGGGAACAGCGCGCAGCUGUCAACGCGCCGGAAACUAUGCAGCGAA--UUUCCGUUUCCGGCGCCCGUUUCACAGAGCAUGUCUUUUCCCGAUGCAA ----------.......(((((((((.((((.((..((((.((((((((((((((...........--.....)))))))))))..)))..)))).)).)).)).)))))))))... ( -42.49, z-score = -4.13, R) >droSim1.chr2L 4405312 105 - 22036055 ----------CAUUCUCCAUCGGGAAGAGCGCGCAGCUGUCAACGCGCCGGAAACUAUGCAGCGAA--UUUCCGUUUCCGGCGCCCGUUUCACAGAGCAUGUCUUUUCCCAAUGCGA ----------.....(((((.((((((((((.((..((((.((((((((((((((...........--.....)))))))))))..)))..)))).)).)).)))))))).))).)) ( -40.99, z-score = -3.30, R) >dp4.chr4_group3 322230 106 + 11692001 --------UAUUUCCCCCAGCAAGGAGAGUCCGCUGCUGUCAACGCACCGGAAACUACGCAGCGAA--UUUUCGUUUCCGGUGCCUGUUC-AUAGAGCGCCACUGUACUUGAUGGGA --------.......((((.((((.(.(((.((((.((((.((((((((((((((...........--.....)))))))))))..))).-))))))))..))).).)))).)))). ( -41.49, z-score = -3.60, R) >droPer1.super_8 1363852 106 + 3966273 --------UAUUUCCCCCAGCAAGGAGAGUCCGCUGCUGUCAACGCACCGGAAACUACGCAGCGAA--UUUUCGUUUCCGGUGCCUGUUC-AUACAGCGCCACUGUACUUGAUGGGA --------.......((((.((((.(.(((.(((((.(((.((((((((((((((...........--.....)))))))))))..))).-))))))))..))).).)))).)))). ( -41.29, z-score = -3.74, R) >droAna3.scaffold_12943 1678168 104 + 5039921 ----------CUUUUGCCAUCCGGAAGAGGGCGCUGCUGUCAACGCGCCGGAAACUAUGCAGCGAA--UUUUCGUUUCCGGCGCCCGUUU-ACGCAGCGCUUCUGUCCCUGAUGCGA ----------...((((.(((.((.(.((((((((((.((.((((((((((((((...........--.....)))))))))))..))).-))))))))).))).).)).))))))) ( -46.89, z-score = -3.56, R) >droWil1.scaffold_180703 1222562 111 + 3946847 CAGGAGAGAAAAUUUCUAGUCCAGUUAAGGCCA-AGCUGUCAACGCUCCGAAAACUAUGCAGCGAAUUUUUUAGUUUUCUCAGCCUGUUC-ACAGAAUGAUCUUGAUCUGCGA---- ..(((((.....)))))..((((((((((..((-..((((.((((((..((((((((..............))))))))..)))..))).-))))..))..))))).))).))---- ( -28.24, z-score = -0.92, R) >droVir3.scaffold_12963 8392800 95 + 20206255 ---------GCGCAUCCAUCCAAGCACAGAACGCAGCUGUUAACGCGCCUGAAAUUAUGCAGCGAA-UUUUUUGUUUCUGGCGCCUGUUU-ACAGCGCGCAACUGU----------- ---------((((..........((.......)).(((((.((((((((.(((((...........-......))))).)))))..))).-)))))))))......----------- ( -30.83, z-score = -1.34, R) >droMoj3.scaffold_6500 11836437 94 + 32352404 ----------CGCAGCCAGCCAAGCGCAGAGCGCAGCUGUUAACGCGCUUGAAGCUAUGCAGCGAA-AUUUUUGCUUCUCGCGCCUGUUC-ACAGUGCACCACUGU----------- ----------.((((.((((...((((...)))).)))).....((((..(((((...........-......)))))..))))))))..-((((((...))))))----------- ( -30.33, z-score = 0.59, R) >droGri2.scaffold_15126 3030152 93 + 8399593 ----------CGCUUUCGAUCAAGCACAAAGCG-AGCUGUUAACGCGCCUGAAGCUGUGCAGCGAA-UUUUUGGCUUCUCGCGCCUGUUC-AUAGAGCGUAGUUGU----------- ----------.((((......))))((((.(((-..((((.(((((((..(((((((.........-....)))))))..))))..))).-))))..)))..))))----------- ( -27.82, z-score = 0.60, R) >consensus __________CAUUCUCCAUCAAGAACAGCGCGCAGCUGUCAACGCGCCGGAAACUAUGCAGCGAA__UUUUCGUUUCCGGCGCCCGUUU_ACAGAGCAUGUCUGUUCCCGAUGCGA ....................................((((..((((((..(((((..................)))))..))))..))...))))...................... (-11.12 = -10.14 + -0.98)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:34 2011