
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,829,750 – 6,829,854 |
| Length | 104 |
| Max. P | 0.527121 |

| Location | 6,829,750 – 6,829,854 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.91 |
| Shannon entropy | 0.52329 |
| G+C content | 0.49899 |
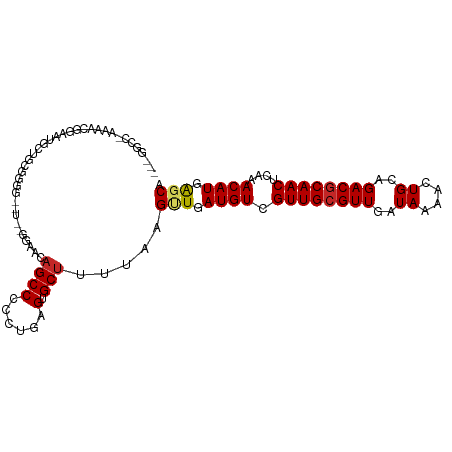
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -15.01 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
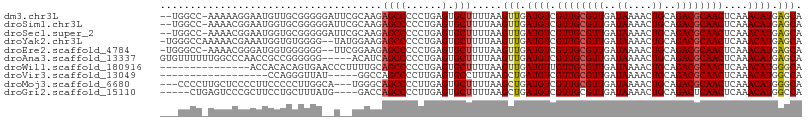
>dm3.chr3L 6829750 104 - 24543557 --UGGCC-AAAAAGGAAUGUUGCGGGGGAUUCGCAAGAGCCCCCUGAGUGCUUUUAAGUUGAUGUCGUUGCGUUGAUAAAACUGCAGACGCAACUCAAACAUGAGCA --.....-..(((((.((....((((((..((....))..)))))).)).)))))..(((.((((.((((((((..((....))..))))))))....)))).))). ( -34.20, z-score = -1.91, R) >droSim1.chr3L 6311405 104 - 22553184 --UGGCC-AAAACGGAAUGGUGCGGGGGAUUCGCAAGAGCCCCCUGAGUGCUUUUAAGUUGAUGUCGUUGCGUUGAUAAAACUGCAGACGCAACUCAAACAUGAGCA --..(((-(........))))(((((((.(((....))))))))(((((((.(((.((((..(((((......))))).))))..))).)).))))).......)). ( -35.80, z-score = -2.00, R) >droSec1.super_2 6763952 104 - 7591821 --UGGCC-AAAACGGAAUGGUGCGGGGGAUUCGCAAGAGCCCCCUGAGUGCUUUUAAGUUGAUGUCGUUGCGUUGAUAAAACUGCAGACGCAACUCAAACAUGAGCA --..(((-(........))))(((((((.(((....))))))))(((((((.(((.((((..(((((......))))).))))..))).)).))))).......)). ( -35.80, z-score = -2.00, R) >droYak2.chr3L 7418143 104 - 24197627 -UGGGCCAAAAACGAAAUGGUGUGGGG--UAUGGAAGAGCCCCCUGAGUGCUUUUAAGUUGAUGUCGUUGCGUUGAUAAAACUGCAGACGCAACUCAAACAUGAGCA -.(.((((.........)))).)((((--(........))))).....(((((.....((((....((((((((..((....))..))))))))))))....))))) ( -27.00, z-score = -0.18, R) >droEre2.scaffold_4784 21232951 103 + 25762168 -UGGGCC-AAAACGGGAUGGUGGGGGG--UUCGGAAGAGCCCCCUGAGUGCUUUUAAGUUGAUGUCGUUGCGUUGAUAAAACUGCAGACGCAACUCAAACAUGAGCA -...(((-(........))))((((((--(((....)))))))))...(((((.....((((....((((((((..((....))..))))))))))))....))))) ( -40.30, z-score = -3.04, R) >droAna3.scaffold_13337 3215059 102 + 23293914 GUGUUUUUUGGCCCAACCGCCGGGGGG-----ACAUCAGCCCCCUGAGUGCUUUUAAGUUGAUGUCGUUGCGUUGAUAAAACUGCAGACGCAACUCAAACAUGAGCA ((((((...(((.((((((((((((((-----.(....)))))))).))).......))))..)))((((((((..((....))..))))))))..))))))..... ( -33.11, z-score = -1.34, R) >droWil1.scaffold_180916 2376889 92 + 2700594 ---------------ACCACACAGUGAACCCUUUUGCAGCCCCCUGAGUGCUUUUAAGUUGAUGUUGUUGCGUUGAUAAAACUGCAGACGCAACUCAAACAUGGGCA ---------------........(..((....))..).((((..(((((........((..((((....))))..)).....(((....)))))))).....)))). ( -20.60, z-score = 0.27, R) >droVir3.scaffold_13049 7183142 84 - 25233164 ------------------CCAGGGUUAU-----GGCCAGCCCCUUGAGUGCCUUUAAGCUGAUGUCGUUGCGUUGAUAAAACUGCAGACGCAACUCAAACAUGGCCA ------------------....((((((-----(..((((...(((((....))))))))).....((((((((..((....))..)))))))).....))))))). ( -23.60, z-score = -0.40, R) >droMoj3.scaffold_6680 5451433 101 - 24764193 ---CCCCUUGCUCCCCUUCCCCCUUGGCA---UGGGCAGCCCCUUGAGUGCUUUUAAGCUGAUGUCGUUGCGUUGAUAAAACUGCAGACGCAACUCAAACAUGGGCA ---.(((.((((.............))))---.)))..((((.(((((((((....)))...((.(((((((((.....))).))).)))))))))))....)))). ( -28.92, z-score = -0.67, R) >droGri2.scaffold_15110 18888098 98 + 24565398 -----CUGAGUCCCGCUUCCUGCUUUAUG----GACCAGCCCCUUGAGUGCUUUUAAGCUGAUGUCGUUGCGUUGAUAAAACUGCAGACUCAACUCAAACAUGGCCA -----.((.((((.((.....)).....)----)))))(((..(((((((((....)))(((.(((..((((((.....))).)))))))))))))))....))).. ( -24.20, z-score = -0.95, R) >consensus ___GGCC_AAAACGGAAUGCUGCGGGG__U__GGAACAGCCCCCUGAGUGCUUUUAAGUUGAUGUCGUUGCGUUGAUAAAACUGCAGACGCAACUCAAACAUGAGCA .....................................((((......).))).....(((.((((.((((((((..((....))..))))))))....)))).))). (-15.01 = -15.10 + 0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:12 2011