
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,828,550 – 6,828,644 |
| Length | 94 |
| Max. P | 0.878294 |

| Location | 6,828,550 – 6,828,644 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 88.83 |
| Shannon entropy | 0.18218 |
| G+C content | 0.34683 |
| Mean single sequence MFE | -16.30 |
| Consensus MFE | -15.22 |
| Energy contribution | -14.74 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.878294 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
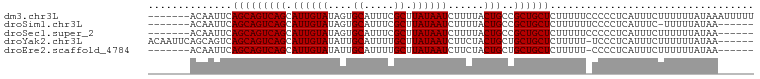
>dm3.chr3L 6828550 94 + 24543557 AAAAAUUUAUAAAAAAGAAAUGAGGGGGAAAAAGAGCAGCGGCAGUAAAAGAUUAUAAGCGAAAUGCACUAUACAAUGCUGACUGCUGAAUUGU------- ..................................((((((((((((((....))))..((.....)).........))))).))))).......------- ( -14.50, z-score = -0.78, R) >droSim1.chr3L 6310229 87 + 22553184 ------UUAUAAAAA-GAAAUGAGGGGAAAAAAGAGCAGCGGCAGUAAAAGAUUAUAAGCGAAAUGCACUAUACAAUGCUGACUGCUGAAUUGU------- ------.........-..................((((((((((((((....))))..((.....)).........))))).))))).......------- ( -14.50, z-score = -1.01, R) >droSec1.super_2 6762793 88 + 7591821 ------UUAUAAAAAAGAAAUGAGGGGGAAAAAGAGCAGCGGCAGUAAAAGAUUAUAAGCGAAAUGCACUAUACAAUGCUGACUGCUGAAUUGU------- ------............................((((((((((((((....))))..((.....)).........))))).))))).......------- ( -14.50, z-score = -0.98, R) >droYak2.chr3L 7416943 94 + 24197627 ------UUAUAAAAAAGAAAUGAGGGA-AAAAAGAGCAGCAGCAGUAGAAGAUUAUAAGCAAAAUGCAAUAUACAAUGCUGACUGCUGACUGCUGAAUUGU ------.....................-......((((((((((((((.....((((.((.....))..)))).....)).))))))).)))))....... ( -21.50, z-score = -2.68, R) >droEre2.scaffold_4784 21231770 87 - 25762168 ------UUAUAAAAAAGAAAUGAGGGG-AAAAAGAGCAGCAGCAGUAGAAGAUUAUAAGCAAAAUGCAAUAUACAAUGCUGACUGCUGAAUUGU------- ------.....................-........(((((((((((......(((..((.....)).))).....))))).))))))......------- ( -16.50, z-score = -2.01, R) >consensus ______UUAUAAAAAAGAAAUGAGGGG_AAAAAGAGCAGCGGCAGUAAAAGAUUAUAAGCGAAAUGCACUAUACAAUGCUGACUGCUGAAUUGU_______ ..................................((((((((((((((....))))..((.....)).........))))).))))).............. (-15.22 = -14.74 + -0.48)

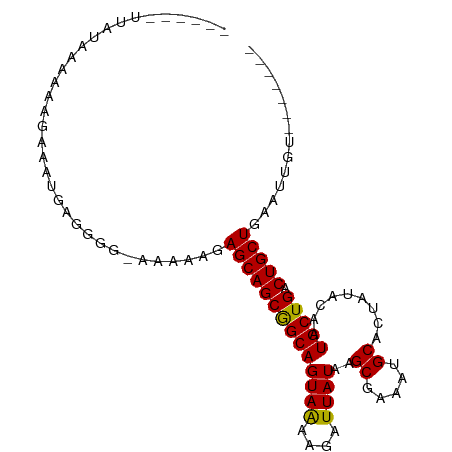

| Location | 6,828,550 – 6,828,644 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 88.83 |
| Shannon entropy | 0.18218 |
| G+C content | 0.34683 |
| Mean single sequence MFE | -14.36 |
| Consensus MFE | -10.12 |
| Energy contribution | -10.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.754475 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 6828550 94 - 24543557 -------ACAAUUCAGCAGUCAGCAUUGUAUAGUGCAUUUCGCUUAUAAUCUUUUACUGCCGCUGCUCUUUUUCCCCCUCAUUUCUUUUUUAUAAAUUUUU -------.......((((((..((((((((.((((.....)))))))))........))).)))))).................................. ( -11.50, z-score = -1.55, R) >droSim1.chr3L 6310229 87 - 22553184 -------ACAAUUCAGCAGUCAGCAUUGUAUAGUGCAUUUCGCUUAUAAUCUUUUACUGCCGCUGCUCUUUUUUCCCCUCAUUUC-UUUUUAUAA------ -------.......((((((..((((((((.((((.....)))))))))........))).))))))..................-.........------ ( -11.50, z-score = -1.44, R) >droSec1.super_2 6762793 88 - 7591821 -------ACAAUUCAGCAGUCAGCAUUGUAUAGUGCAUUUCGCUUAUAAUCUUUUACUGCCGCUGCUCUUUUUCCCCCUCAUUUCUUUUUUAUAA------ -------.......((((((..((((((((.((((.....)))))))))........))).))))))............................------ ( -11.50, z-score = -1.73, R) >droYak2.chr3L 7416943 94 - 24197627 ACAAUUCAGCAGUCAGCAGUCAGCAUUGUAUAUUGCAUUUUGCUUAUAAUCUUCUACUGCUGCUGCUCUUUUU-UCCCUCAUUUCUUUUUUAUAA------ .......(((((.(((((((.((.((((((....((.....)).))))))))...))))))))))))......-.....................------ ( -22.50, z-score = -4.31, R) >droEre2.scaffold_4784 21231770 87 + 25762168 -------ACAAUUCAGCAGUCAGCAUUGUAUAUUGCAUUUUGCUUAUAAUCUUCUACUGCUGCUGCUCUUUUU-CCCCUCAUUUCUUUUUUAUAA------ -------.......(((((.((((((((((....((.....)).)))))........))))))))))......-.....................------ ( -14.80, z-score = -2.59, R) >consensus _______ACAAUUCAGCAGUCAGCAUUGUAUAGUGCAUUUCGCUUAUAAUCUUUUACUGCCGCUGCUCUUUUU_CCCCUCAUUUCUUUUUUAUAA______ ..............(((((((((.((((((....((.....)).))))))......)))..)))))).................................. (-10.12 = -10.12 + 0.00)
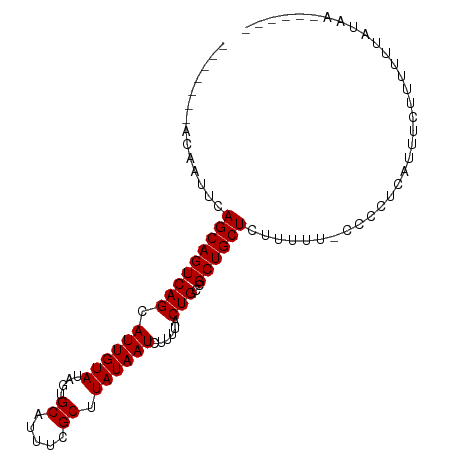
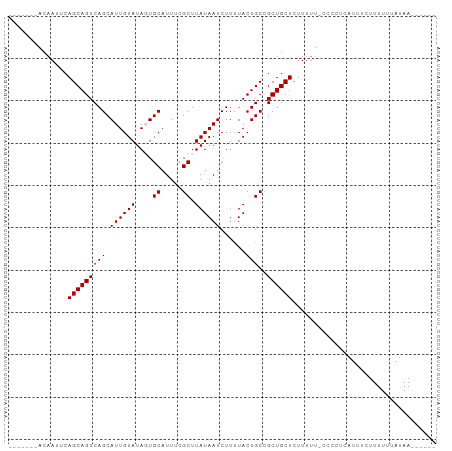
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:12 2011