
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,823,157 – 6,823,255 |
| Length | 98 |
| Max. P | 0.995211 |

| Location | 6,823,157 – 6,823,247 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 83.49 |
| Shannon entropy | 0.30988 |
| G+C content | 0.45911 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -16.99 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6823157 90 + 24543557 ---------------CCUGGUCGGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAUCAU- ---------------..((((..(((....(((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))))).)))...)))).- ( -28.10, z-score = -2.96, R) >droSim1.chr3L 6304870 90 + 22553184 ---------------CCUGGUCGGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAUCAU- ---------------..((((..(((....(((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))))).)))...)))).- ( -28.10, z-score = -2.96, R) >droSec1.super_2 6757465 90 + 7591821 ---------------CCUGGUCGGACUUACAACAUUGCCCACAAUUUCCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAUCAU- ---------------..((((..(((....(((((.((((((((((...((.....((((.....)----------)))...)).))))))..)))).))))).)))...)))).- ( -23.90, z-score = -1.92, R) >droYak2.chr3L 7411045 90 + 24197627 ---------------CCUGGUCGGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAUCAU- ---------------..((((..(((....(((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))))).)))...)))).- ( -28.10, z-score = -2.96, R) >droEre2.scaffold_4784 21226058 90 - 25762168 ---------------CCUGGUCGGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAUCAU- ---------------..((((..(((....(((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))))).)))...)))).- ( -28.10, z-score = -2.96, R) >droAna3.scaffold_13337 3207616 90 - 23293914 ---------------CCUGGCCGAACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAUCAU- ---------------.......((....(((((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))).)))).....))..- ( -24.10, z-score = -1.94, R) >dp4.chrXR_group8 4118107 100 - 9212921 ---------------CAUCUACCGACUUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUCGAGUCGAGUCGAGAGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCCUCCU- ---------------........(((....((((((((((((((((((((((....(((((......)))))..)))))).....))))))..)))))))))).)))........- ( -38.20, z-score = -5.53, R) >droPer1.super_38 597034 100 + 801819 ---------------CAUCUACCGACUUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUCGAGUCGAGUCGAGAGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCCUCCU- ---------------........(((....((((((((((((((((((((((....(((((......)))))..)))))).....))))))..)))))))))).)))........- ( -38.20, z-score = -5.53, R) >droWil1.scaffold_180916 335251 94 + 2700594 ----------CUUCAACUCUUCG-ACUUACAAUAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAUCGU- ----------............(-((....(((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))))).)))........- ( -24.90, z-score = -3.02, R) >droVir3.scaffold_13049 7175448 90 + 25233164 --------------ACUAACC-AGACUUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUUGAG----------AGCGUUGACAAUUGUUUGGGCCUUGUUUGUCGACGUCAA- --------------.......-.(((((((((((.(((((((((((.(.((..(((((((.....)----------)))))))))))))))..))))).)).)))).)).)))..- ( -30.90, z-score = -3.78, R) >droMoj3.scaffold_6680 5444008 90 + 24764193 --------------ACUUACA--GACCUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUUGAG----------AGCGUUGACAAUUGUUCGGGCCUUGUUUGUCGACGUUCAG --------------.......--(((..((((...(((((((((((.(.((..(((((((.....)----------)))))))))))))))..)))))))))..)))......... ( -29.80, z-score = -3.38, R) >droGri2.scaffold_15110 18880789 105 - 24565398 UUGCGACAACGACUACUAACAGAGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCGUUGACAAUUGUUUGGGCCUUGUUUGUCGACAUCGA- .(((((((.......((.....))......((((..((((((((((.(.((..(((((((.....)----------)))))))))))))))..))))..))))))))).))....- ( -29.50, z-score = -2.29, R) >consensus _______________CCUGGUCGGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG__________AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAUCAU_ .......................(((....((((..((((((((((.((.......))..(((((...............)))))))))))..))))..)))).)))......... (-16.99 = -17.03 + 0.04)



| Location | 6,823,157 – 6,823,247 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.49 |
| Shannon entropy | 0.30988 |
| G+C content | 0.45911 |
| Mean single sequence MFE | -29.91 |
| Consensus MFE | -18.13 |
| Energy contribution | -18.04 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6823157 90 - 24543557 -AUGAUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG--------------- -.....(((....(((((.((((..((((((.....((((----------((.(((....))).)))))))))))))))).)))))....)))........--------------- ( -27.50, z-score = -2.18, R) >droSim1.chr3L 6304870 90 - 22553184 -AUGAUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG--------------- -.....(((....(((((.((((..((((((.....((((----------((.(((....))).)))))))))))))))).)))))....)))........--------------- ( -27.50, z-score = -2.18, R) >droSec1.super_2 6757465 90 - 7591821 -AUGAUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGGAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG--------------- -.....(((....(((((.(((((.....((.((((((((----------(.....)))))))..)).)).....))))).)))))....)))........--------------- ( -25.80, z-score = -1.64, R) >droYak2.chr3L 7411045 90 - 24197627 -AUGAUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG--------------- -.....(((....(((((.((((..((((((.....((((----------((.(((....))).)))))))))))))))).)))))....)))........--------------- ( -27.50, z-score = -2.18, R) >droEre2.scaffold_4784 21226058 90 + 25762168 -AUGAUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG--------------- -.....(((....(((((.((((..((((((.....((((----------((.(((....))).)))))))))))))))).)))))....)))........--------------- ( -27.50, z-score = -2.18, R) >droAna3.scaffold_13337 3207616 90 + 23293914 -AUGAUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUUCGGCCAGG--------------- -.((.(((..((((.(((.((((..((((((.....((((----------((.(((....))).)))))))))))))))).)))))))....)))..))..--------------- ( -27.40, z-score = -1.81, R) >dp4.chrXR_group8 4118107 100 + 9212921 -AGGAGGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCUCUCGACUCGACUCGACGAGCGUUUAGAGCAAAUUGUGGGCCAUGUUGUAAGUCGGUAGAUG--------------- -.....(((....((((((((((..((((((((.(((((((((((......)))).)))))))....)).))))))))))))))))....)))........--------------- ( -37.60, z-score = -3.81, R) >droPer1.super_38 597034 100 - 801819 -AGGAGGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCUCUCGACUCGACUCGACGAGCGUUUAGAGCAAAUUGUGGGCCAUGUUGUAAGUCGGUAGAUG--------------- -.....(((....((((((((((..((((((((.(((((((((((......)))).)))))))....)).))))))))))))))))....)))........--------------- ( -37.60, z-score = -3.81, R) >droWil1.scaffold_180916 335251 94 - 2700594 -ACGAUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUAUUGUAAGU-CGAAGAGUUGAAG---------- -.((((....((((.....((((..((((((.....((((----------((.(((....))).))))))))))))))))....))))..))-))...........---------- ( -27.50, z-score = -2.58, R) >droVir3.scaffold_13049 7175448 90 - 25233164 -UUGACGUCGACAAACAAGGCCCAAACAAUUGUCAACGCU----------CUCAACGAGCGUUUAGAGCAAAUUGUGGGCCAUGUUGUAAGUCU-GGUUAGU-------------- -..(((....((((.((.(((((..((((((((.((((((----------(.....)))))))....)).))))))))))).))))))..))).-.......-------------- ( -31.40, z-score = -3.43, R) >droMoj3.scaffold_6680 5444008 90 - 24764193 CUGAACGUCGACAAACAAGGCCCGAACAAUUGUCAACGCU----------CUCAACGAGCGUUUAGAGCAAAUUGUGGGCCAUGUUGUAGGUC--UGUAAGU-------------- .........(((.((((.(((((..((((((((.((((((----------(.....)))))))....)).))))))))))).))))....)))--.......-------------- ( -30.90, z-score = -3.02, R) >droGri2.scaffold_15110 18880789 105 + 24565398 -UCGAUGUCGACAAACAAGGCCCAAACAAUUGUCAACGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCUCUGUUAGUAGUCGUUGUCGCAA -.(((((.((((.((((..((((..((((((((.((((((----------(.....)))))))....)).))))))))))..))))......((.....))..)))).)))))... ( -30.70, z-score = -1.62, R) >consensus _AUGAUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU__________CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG_______________ .........(((.((((..((((..((((((.((((((((.................))))))..))...))))))))))..))))....)))....................... (-18.13 = -18.04 + -0.09)


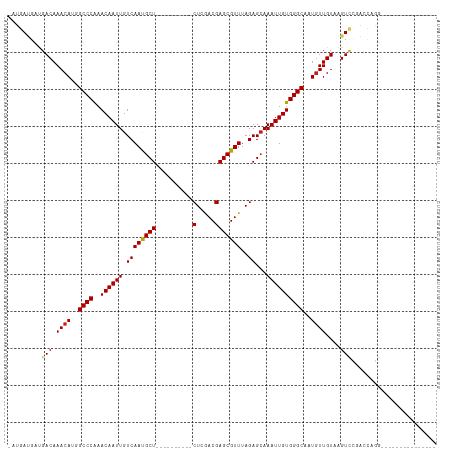
| Location | 6,823,157 – 6,823,255 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Shannon entropy | 0.36762 |
| G+C content | 0.47194 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -17.50 |
| Energy contribution | -17.45 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
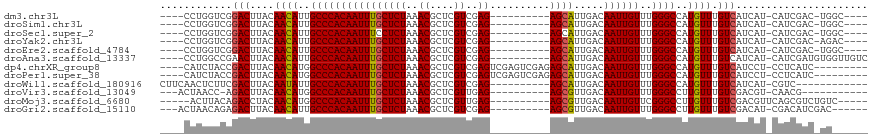
>dm3.chr3L 6823157 98 + 24543557 ----CCUGGUCGGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAU-CAUCGAC-UGGC---- ----.(..(((((((....(((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))))).))).....-...))))-..).---- ( -32.00, z-score = -2.96, R) >droSim1.chr3L 6304870 98 + 22553184 ----CCUGGUCGGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAU-CAUCGAC-UGGC---- ----.(..(((((((....(((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))))).))).....-...))))-..).---- ( -32.00, z-score = -2.96, R) >droSec1.super_2 6757465 98 + 7591821 ----CCUGGUCGGACUUACAACAUUGCCCACAAUUUCCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAU-CAUCGAC-UGGC---- ----.(..(((((((....(((((.((((((((((...((.....((((.....)----------)))...)).))))))..)))).))))).))).....-...))))-..).---- ( -27.80, z-score = -1.92, R) >droYak2.chr3L 7411045 98 + 24197627 ----CCUGGUCGGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAU-CAUCGAC-AGAC---- ----.(((.((((((....(((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))))).))).....-...))))-))..---- ( -28.90, z-score = -2.35, R) >droEre2.scaffold_4784 21226058 98 - 25762168 ----CCUGGUCGGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAU-CAUCGAC-UGGC---- ----.(..(((((((....(((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))))).))).....-...))))-..).---- ( -32.00, z-score = -2.96, R) >droAna3.scaffold_13337 3207616 103 - 23293914 ----CCUGGCCGAACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAU-CAUCGAUGUGGUUGUC ----...(((((.....(((((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))).))))((((..-....)))))))))... ( -30.20, z-score = -1.86, R) >dp4.chrXR_group8 4118107 104 - 9212921 ----CAUCUACCGACUUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUCGAGUCGAGUCGAGAGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCCU-CCUCAUC--------- ----........(((....((((((((((((((((((((((....(((((......)))))..)))))).....))))))..)))))))))).))).....-.......--------- ( -38.20, z-score = -5.40, R) >droPer1.super_38 597034 104 + 801819 ----CAUCUACCGACUUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUCGAGUCGAGUCGAGAGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCCU-CCUCAUC--------- ----........(((....((((((((((((((((((((((....(((((......)))))..)))))).....))))))..)))))))))).))).....-.......--------- ( -38.20, z-score = -5.40, R) >droWil1.scaffold_180916 335251 95 + 2700594 CUUCAACUCUUCGACUUACAAUAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAU-CGUC------------ ............(((....(((((.((((((((((((((((..((....))..))----------)))).....))))))..)))).))))).))).....-....------------ ( -24.90, z-score = -3.08, R) >droVir3.scaffold_13049 7175448 92 + 25233164 ---ACUAACC-AGACUUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUUGAG----------AGCGUUGACAAUUGUUUGGGCCUUGUUUGUCGACGU-CAACG----------- ---.......-.(((((((((((.(((((((((((.(.((..(((((((.....)----------)))))))))))))))..))))).)).)))).)).))-)....----------- ( -30.90, z-score = -3.72, R) >droMoj3.scaffold_6680 5444008 98 + 24764193 -----ACUUACAGACCUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUUGAG----------AGCGUUGACAAUUGUUCGGGCCUUGUUUGUCGACGUUCAGCGUCUGUC----- -----.......(((..((((...(((((((((((.(.((..(((((((.....)----------)))))))))))))))..)))))))))..)))((((.....))))....----- ( -34.90, z-score = -3.57, R) >droGri2.scaffold_15110 18880803 98 - 24565398 ---ACUAACAGAGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG----------AGCGUUGACAAUUGUUUGGGCCUUGUUUGUCGACAU-CGACAUCGAC------ ---.......((.......((((..((((((((((.(.((..(((((((.....)----------)))))))))))))))..))))..))))(((((....-)))))))...------ ( -30.00, z-score = -3.10, R) >consensus ____CCUGGUCGGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUCGAG__________AGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAU_CAUCGAC_UG______ ............(((....((((..((((((((((.((.......))..(((((...............)))))))))))..))))..)))).)))...................... (-17.50 = -17.45 + -0.05)


| Location | 6,823,157 – 6,823,255 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.38 |
| Shannon entropy | 0.36762 |
| G+C content | 0.47194 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.52 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6823157 98 - 24543557 ----GCCA-GUCGAUG-AUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG---- ----.((.-((((..(-((....((((.(((.((((..((((((.....((((----------((.(((....))).)))))))))))))))).)))))))..)))))))..))---- ( -32.10, z-score = -2.57, R) >droSim1.chr3L 6304870 98 - 22553184 ----GCCA-GUCGAUG-AUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG---- ----.((.-((((..(-((....((((.(((.((((..((((((.....((((----------((.(((....))).)))))))))))))))).)))))))..)))))))..))---- ( -32.10, z-score = -2.57, R) >droSec1.super_2 6757465 98 - 7591821 ----GCCA-GUCGAUG-AUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGGAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG---- ----.((.-((((..(-((....((((.(((.(((((.....((.((((((((----------(.....)))))))..)).)).....))))).)))))))..)))))))..))---- ( -30.40, z-score = -1.98, R) >droYak2.chr3L 7411045 98 - 24197627 ----GUCU-GUCGAUG-AUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG---- ----..((-((((..(-((....((((.(((.((((..((((((.....((((----------((.(((....))).)))))))))))))))).)))))))..)))))).))).---- ( -32.50, z-score = -2.66, R) >droEre2.scaffold_4784 21226058 98 + 25762168 ----GCCA-GUCGAUG-AUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG---- ----.((.-((((..(-((....((((.(((.((((..((((((.....((((----------((.(((....))).)))))))))))))))).)))))))..)))))))..))---- ( -32.10, z-score = -2.57, R) >droAna3.scaffold_13337 3207616 103 + 23293914 GACAACCACAUCGAUG-AUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUUCGGCCAGG---- .....((.((((....-..))))((((.(((.((((..((((((.....((((----------((.(((....))).)))))))))))))))).)))))))......)).....---- ( -30.00, z-score = -1.56, R) >dp4.chrXR_group8 4118107 104 + 9212921 ---------GAUGAGG-AGGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCUCUCGACUCGACUCGACGAGCGUUUAGAGCAAAUUGUGGGCCAUGUUGUAAGUCGGUAGAUG---- ---------.......-..(((....((((((((((..((((((((.(((((((((((......)))).)))))))....)).))))))))))))))))....)))........---- ( -37.60, z-score = -3.45, R) >droPer1.super_38 597034 104 - 801819 ---------GAUGAGG-AGGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCUCUCGACUCGACUCGACGAGCGUUUAGAGCAAAUUGUGGGCCAUGUUGUAAGUCGGUAGAUG---- ---------.......-..(((....((((((((((..((((((((.(((((((((((......)))).)))))))....)).))))))))))))))))....)))........---- ( -37.60, z-score = -3.45, R) >droWil1.scaffold_180916 335251 95 - 2700594 ------------GACG-AUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUAUUGUAAGUCGAAGAGUUGAAG ------------..((-((....((((.....((((..((((((.....((((----------((.(((....))).))))))))))))))))....))))..))))........... ( -27.50, z-score = -2.55, R) >droVir3.scaffold_13049 7175448 92 - 25233164 -----------CGUUG-ACGUCGACAAACAAGGCCCAAACAAUUGUCAACGCU----------CUCAACGAGCGUUUAGAGCAAAUUGUGGGCCAUGUUGUAAGUCU-GGUUAGU--- -----------....(-((....((((.((.(((((..((((((((.((((((----------(.....)))))))....)).))))))))))).))))))..))).-.......--- ( -31.40, z-score = -3.09, R) >droMoj3.scaffold_6680 5444008 98 - 24764193 -----GACAGACGCUGAACGUCGACAAACAAGGCCCGAACAAUUGUCAACGCU----------CUCAACGAGCGUUUAGAGCAAAUUGUGGGCCAUGUUGUAGGUCUGUAAGU----- -----.((((((.(((..........((((.(((((..((((((((.((((((----------(.....)))))))....)).))))))))))).)))).)))))))))....----- ( -36.00, z-score = -3.36, R) >droGri2.scaffold_15110 18880803 98 + 24565398 ------GUCGAUGUCG-AUGUCGACAAACAAGGCCCAAACAAUUGUCAACGCU----------CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCUCUGUUAGU--- ------((((((....-..)))))).((((..((((..((((((((.((((((----------(.....)))))))....)).))))))))))..))))................--- ( -31.70, z-score = -2.57, R) >consensus ______CA_GUCGAUG_AUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCU__________CUCGACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCCGACCAGG____ ......................(((.((((..((((..((((((.((((((((.................))))))..))...))))))))))..))))....)))............ (-18.70 = -18.52 + -0.18)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:09 2011