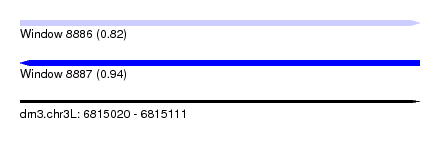
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,815,020 – 6,815,111 |
| Length | 91 |
| Max. P | 0.936655 |

| Location | 6,815,020 – 6,815,111 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.85 |
| Shannon entropy | 0.35082 |
| G+C content | 0.38892 |
| Mean single sequence MFE | -20.36 |
| Consensus MFE | -13.50 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.823664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
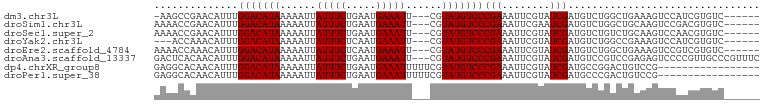
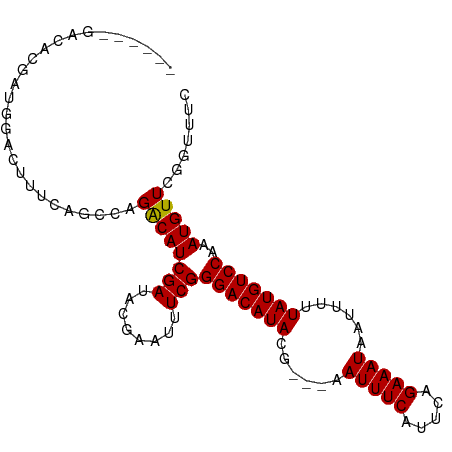
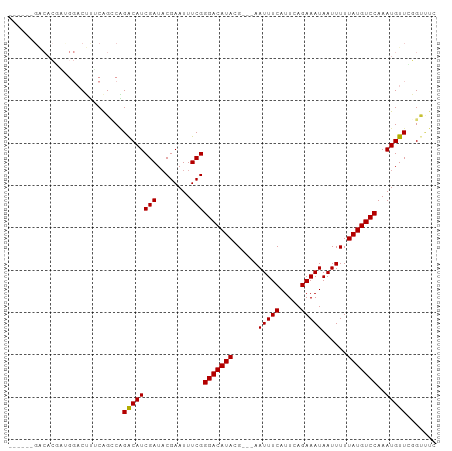
>dm3.chr3L 6815020 91 + 24543557 -AAGCCGAACAUUUGGACAUAAAAAUUAUUUCUGAAUGAAAUU---CGUAUGUCCCGAAAUUCGUAUCGAUGUCUGGCUGAAAGUCCAUCGUGUC------ -.(((((.(((((.(((((((......(((((.....))))).---..)))))))((.....))....))))).)))))................------ ( -22.20, z-score = -2.20, R) >droSim1.chr3L 6296539 92 + 22553184 AAAACCGAACAUUUGGACAUAAAAAUUAUUUCUGAAUGAAAUU---CGUAUGUCCCGAAAUUCGAAUCGAUGUCUGGCUGCAAGUCCGACGUGUC------ .....((((..((.(((((((......(((((.....))))).---..))))))).))..)))).....(((((.(((.....))).)))))...------ ( -23.10, z-score = -2.24, R) >droSec1.super_2 6749166 92 + 7591821 AAAACCGAACAUUUGGACAUAAAAAUUAUUUCUGAAUGAAAUU---CGUAUGUCCCGAAAUUCGUAUCGAUGUCUGUCUGCAAGUCCAACGUGUC------ ........((((((((((...............(((((((.((---((.......)))).))))).))(((....))).....)))))).)))).------ ( -17.40, z-score = -1.06, R) >droYak2.chr3L 7402425 89 + 24197627 ---ACCAAACAUUUGGACAUAAAAAUUAUUUCUCAAUGAAAUU---CGUAUGUCCCGAAAUUCGUAUCGAUGUCUGGCCGAAAGUCCAUCGUGUC------ ---.(((.(((((.(((((((......(((((.....))))).---..)))))))((.....))....))))).))).(....)...........------ ( -18.30, z-score = -1.40, R) >droEre2.scaffold_4784 21217646 92 - 25762168 AAAACCAAACAUUUGGACAUAAAAAUUAUUUCUCAAUGAAAUU---CGUAUGUCCCGAAAUUCGUAUCGAUGUCUGGCUGAAAGUCCGUCGUGUC------ ....(((.(((((.(((((((......(((((.....))))).---..)))))))((.....))....))))).)))..................------ ( -17.80, z-score = -1.30, R) >droAna3.scaffold_13337 3199267 98 - 23293914 GACUCACAACAUUUGGACAUAAAAAUUAUUUCUGAAUGAAAUU---CGUAUGUCCCGAAAUUCGUAUCGAUGUCCGUCCGAGAGUCCCCGUUGCCCGUUUC (((((.........(((((((......(((((.....))))).---..)))))))......(((...((.....))..))))))))............... ( -20.60, z-score = -1.34, R) >dp4.chrXR_group8 4109210 84 - 9212921 GAGGCACAACAUUUGGACAUAAAAAUUAUUUCUGAAUGAAAUUUUUCGUAUGUCCCGAAAUUCGUAUCGAUGCCGGACUGUCCG----------------- ..((((........(((((((..((..(((((.....)))))..))..)))))))(((........))).))))((.....)).----------------- ( -23.00, z-score = -2.65, R) >droPer1.super_38 588036 84 + 801819 GAGGCACAACAUUUGGACAUAAAAAUUAUUUCUGAAUGAAAUUUUUCGUAUGUCCCGAAAUUCGUAUCGAUGCCCGACUGUCCG----------------- (.((((........(((((((..((..(((((.....)))))..))..)))))))(((........))).))))).........----------------- ( -20.50, z-score = -2.28, R) >consensus AAAACCCAACAUUUGGACAUAAAAAUUAUUUCUGAAUGAAAUU___CGUAUGUCCCGAAAUUCGUAUCGAUGUCUGGCUGAAAGUCCAUCGUGUC______ ..............(((((((......(((((.....)))))......)))))))(((........)))................................ (-13.50 = -13.50 + 0.00)
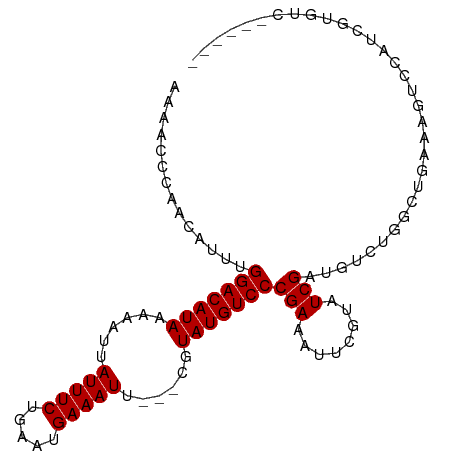
| Location | 6,815,020 – 6,815,111 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.85 |
| Shannon entropy | 0.35082 |
| G+C content | 0.38892 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -15.11 |
| Energy contribution | -14.93 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6815020 91 - 24543557 ------GACACGAUGGACUUUCAGCCAGACAUCGAUACGAAUUUCGGGACAUACG---AAUUUCAUUCAGAAAUAAUUUUUAUGUCCAAAUGUUCGGCUU- ------................((((..(((((((........)))(((((((.(---((((............))))).)))))))..))))..)))).- ( -22.20, z-score = -2.54, R) >droSim1.chr3L 6296539 92 - 22553184 ------GACACGUCGGACUUGCAGCCAGACAUCGAUUCGAAUUUCGGGACAUACG---AAUUUCAUUCAGAAAUAAUUUUUAUGUCCAAAUGUUCGGUUUU ------.....(((((........)).)))......((((((....(((((((.(---((((............))))).)))))))....)))))).... ( -20.90, z-score = -1.26, R) >droSec1.super_2 6749166 92 - 7591821 ------GACACGUUGGACUUGCAGACAGACAUCGAUACGAAUUUCGGGACAUACG---AAUUUCAUUCAGAAAUAAUUUUUAUGUCCAAAUGUUCGGUUUU ------(((.((.((...(((....))).)).))...(((((....(((((((.(---((((............))))).)))))))....)))))))).. ( -17.90, z-score = -0.60, R) >droYak2.chr3L 7402425 89 - 24197627 ------GACACGAUGGACUUUCGGCCAGACAUCGAUACGAAUUUCGGGACAUACG---AAUUUCAUUGAGAAAUAAUUUUUAUGUCCAAAUGUUUGGU--- ------....(((.......)))((((((((((((........)))(((((((.(---((((............))))).)))))))..)))))))))--- ( -24.60, z-score = -2.71, R) >droEre2.scaffold_4784 21217646 92 + 25762168 ------GACACGACGGACUUUCAGCCAGACAUCGAUACGAAUUUCGGGACAUACG---AAUUUCAUUGAGAAAUAAUUUUUAUGUCCAAAUGUUUGGUUUU ------................(((((((((((((........)))(((((((.(---((((............))))).)))))))..)))))))))).. ( -23.40, z-score = -2.76, R) >droAna3.scaffold_13337 3199267 98 + 23293914 GAAACGGGCAACGGGGACUCUCGGACGGACAUCGAUACGAAUUUCGGGACAUACG---AAUUUCAUUCAGAAAUAAUUUUUAUGUCCAAAUGUUGUGAGUC ....(.(....).).(((((.(((.((((..(((...)))..))))(((((((.(---((((............))))).))))))).....))).))))) ( -23.40, z-score = -0.82, R) >dp4.chrXR_group8 4109210 84 + 9212921 -----------------CGGACAGUCCGGCAUCGAUACGAAUUUCGGGACAUACGAAAAAUUUCAUUCAGAAAUAAUUUUUAUGUCCAAAUGUUGUGCCUC -----------------.((.....))((((.(((((((.....))(((((((.(((..(((((.....)))))..))).)))))))...))))))))).. ( -25.00, z-score = -3.31, R) >droPer1.super_38 588036 84 - 801819 -----------------CGGACAGUCGGGCAUCGAUACGAAUUUCGGGACAUACGAAAAAUUUCAUUCAGAAAUAAUUUUUAUGUCCAAAUGUUGUGCCUC -----------------.........(((((.(((((((.....))(((((((.(((..(((((.....)))))..))).)))))))...)))))))))). ( -23.70, z-score = -2.81, R) >consensus ______GACACGAUGGACUUUCAGCCAGACAUCGAUACGAAUUUCGGGACAUACG___AAUUUCAUUCAGAAAUAAUUUUUAUGUCCAAAUGUUCGGUUUC ...........................((((((((........)))(((((((......(((((.....)))))......)))))))..)))))....... (-15.11 = -14.93 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:06 2011