| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,813,196 – 6,813,294 |
| Length | 98 |
| Max. P | 0.875708 |

| Location | 6,813,196 – 6,813,294 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.96 |
| Shannon entropy | 0.34932 |
| G+C content | 0.51663 |
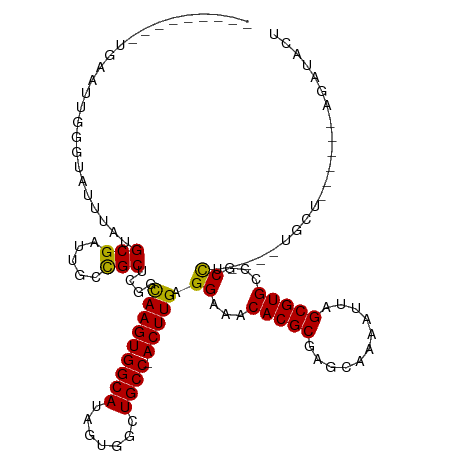
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.58 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
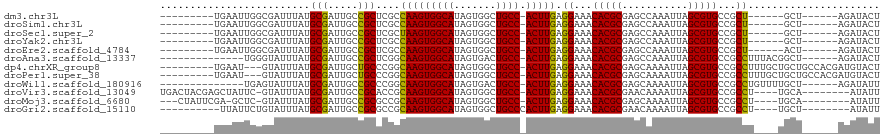
>dm3.chr3L 6813196 98 + 24543557 ---------UGAAUUGGCGAUUUAUGCGAUUGCCGCUCGCCAAGUGGCAUAGUGGCUGCC-ACUUGAGGAAACACGCGAGCCAAAUUAGCGUGCCGCU------GCU------AGAUACU ---------......(((((((.....)))))))(((((((((((((((.......))))-))))).(....)..))))))....((((((......)------)))------))..... ( -39.30, z-score = -2.99, R) >droSim1.chr3L 6294725 98 + 22553184 ---------UGAAUUGGCGAUUUAUGCGAUUGCCGCUCGCCAAGUGGCAUAGUGGCUGCC-ACUUGAGGAAACACGCGAGCCAAAUUAGCGUGCCGCU------GCU------AGAUACU ---------......(((((((.....)))))))(((((((((((((((.......))))-))))).(....)..))))))....((((((......)------)))------))..... ( -39.30, z-score = -2.99, R) >droSec1.super_2 6747379 98 + 7591821 ---------UGAAUUGGCGAUUUAUGCGAUUGCCGCUCGCUAAGUGGCAUAGUGGCUGCC-ACUUGAGGAAACACGCGAGCCAAAUUAGCGUGCCGCU------GCU------AGAUACU ---------......(((((((.....)))))))(((((((((((((((.......))))-))))).(....)..))))))....((((((......)------)))------))..... ( -37.60, z-score = -2.55, R) >droYak2.chr3L 7399910 98 + 24197627 ---------UGAAUUGGCGAUUUAUGCGAUUGCCGCUCGCCAAGUGGCAUAGUGGCUGCC-ACUUGAGGAAACACGCGAGCCAAAUUAGCGUGCCGCU------GCU------AGAUACU ---------......(((((((.....)))))))(((((((((((((((.......))))-))))).(....)..))))))....((((((......)------)))------))..... ( -39.30, z-score = -2.99, R) >droEre2.scaffold_4784 21215847 98 - 25762168 ---------UGAAUUGGCGAUUUAUGCGAUUGCCGCUCGCCAAGUGGCAUAGUGGCUGCC-ACUUGAGGAAACACGCGAGCCAAAUUAGCGUGCCGCU------ACU------AGAUACU ---------......(((((((.....)))))))(((((((((((((((.......))))-))))).(....)..)))))).....(((((...))))------)..------....... ( -38.70, z-score = -3.37, R) >droAna3.scaffold_13337 3197495 99 - 23293914 --------------UGGGUAUUUAUGCGAUUGCCGCUCGGCAAGUGGCAUAGUGACUGCC-ACUUGAGGAAACACGCGAGCCAAAUUAGCGUGCCGCCUUUACGGCU------AGAUACU --------------..(((((((((((((((...(((((.(((((((((.......))))-))))).(....)...)))))..)))).))))((((......)))).------))))))) ( -38.70, z-score = -2.83, R) >dp4.chrXR_group8 4107495 107 - 9212921 ---------UGAAU---GUAUUUAUGCGAUUGCUGCCCGGCAAGUGGCAUAGUGGCUGCC-ACUUGAGGAAACACGCGAGCAAAAUUAGCGUGCCGCCUUUGCUGCUGCCACGAUGUACU ---------.....---((((.(((((.((((((....))).))).)))))(((((.((.-....((((...(((((...........)))))...))))....)).)))))...)))). ( -36.60, z-score = -1.00, R) >droPer1.super_38 586324 107 + 801819 ---------UGAAU---GUAUUUAUGCGAUUGCUGCCCGGCAAGUGGCAUAGUGGCUGCC-ACUUGAGGAAACACGCGAGCAAAAUUAGCGUGCCGCCUUUGCUGCUGCCACGAUGUACU ---------.....---((((.(((((.((((((....))).))).)))))(((((.((.-....((((...(((((...........)))))...))))....)).)))))...)))). ( -36.60, z-score = -1.00, R) >droWil1.scaffold_180916 2353674 99 - 2700594 --------------UGAGUAUUUAUGCGAUUGCCGCCCGGCAAGUGGCAUAGUGACUGCC-ACUUGAGGAAACACGCGAGCAAAAUUAGCGUGCCGCCUGUUUUGCU------AGAUAUU --------------..(((((((.((((...(((....)))((((((((.......))))-))))..(....).))))((((((((..(((...)))..))))))))------))))))) ( -35.20, z-score = -2.77, R) >droVir3.scaffold_13049 7164427 106 + 25233164 UGACUACGAGCUAUUC-GUAUUUAUGCGAUUGCCGCACCGCAAGUGGCAUAGUGGCUGCC-ACUUGAGGAAACACGCGAACAAAAUUAGCGUGCCGCCU----UGCA--------AUAUU (((.((((((...)))-))).)))(((((..((.((.((.(((((((((.......))))-))))).))....((((...........)))))).)).)----))))--------..... ( -34.40, z-score = -1.83, R) >droMoj3.scaffold_6680 5433163 102 + 24764193 ---CUAUUCGA-GCUC-GUAUUUAUGCGAUUGCCGCGCCGCAAGUGGCAUAGUGGCUGCC-ACUUGAGGAAACACGCGAGCAAAAUUAGCGUGCCGCCU----UGCA--------AUAUU ---.(((((((-(.((-(((....)))))((((((((((.(((((((((.......))))-))))).)).....)))).)))).....(((...)))))----)).)--------))).. ( -33.60, z-score = -0.50, R) >droGri2.scaffold_15110 18870346 98 - 24565398 ----------UUAUUCUGUAUUUAUGCGAUUGCCGCGCCGCAAGUGGCAUAGUGGCUGCCCACUUGAGGAAACACGCGAACAAAAUUAGCGUGCCGCCU----UGCU--------AUAUU ----------...............(((.....)))...(((((((((..(((((....)))))...(....)((((...........)))))))).))----))).--------..... ( -27.30, z-score = -0.04, R) >consensus _________UGAAUUGGGUAUUUAUGCGAUUGCCGCUCGGCAAGUGGCAUAGUGGCUGCC_ACUUGAGGAAACACGCGAGCAAAAUUAGCGUGCCGCCU____UGCU______AGAUACU .........................((...((((((.......))))))..(((((...........(....)((((...........))))))))).......)).............. (-22.46 = -22.58 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:04 2011