
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,794,927 – 6,795,040 |
| Length | 113 |
| Max. P | 0.601242 |

| Location | 6,794,927 – 6,795,040 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 125 |
| Reading direction | reverse |
| Mean pairwise identity | 69.27 |
| Shannon entropy | 0.55693 |
| G+C content | 0.46170 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -11.95 |
| Energy contribution | -11.93 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.601242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6794927 113 - 24543557 AGCUGAUCUAUUCAUUUAGGUGCGAGCUGUGACGUCACAGUUU---GUCAAAGGG---AAAUAAAUACAUACAUAUGUAUAUGCAGGGCGGUCAGAAAAGAGGACAAAAUCGGCGAUGU------ .((((((((.(((.(((...(((((((((((....))))))))---).))...))---).....((((((....))))))..............))).)))........))))).....------ ( -25.80, z-score = -0.71, R) >dp4.chrXR_group8 4085945 118 + 9212921 -----AGCCA-CCAAUUGGCUGCAGUUGUUGUUGUCAAAGCCUCUGGUCAGAGAGCCCAAGACUAAAGUCACAUUGAAACGCCCAGGGC-GCCAUAAUCAUAGACGACUACGACUAUGACUACGA -----(((((-.....)))))..((((((.((((((.....((((....)))).((((..(((....)))....((.......))))))-............)))))).)))))).......... ( -34.70, z-score = -1.96, R) >droEre2.scaffold_4784 21197500 99 + 25762168 CGCUGAGGUAUCCAUCUGGAUGCGAGCCGUGACGUCACAGUUC---GUCAUAAGG---AAAUA--------------UAUAUGCAGGGCGGUCAGAAAAGAGGCCAAAAUCGGCGAUGU------ ((((((.((((((....))))))..(((((((((........)---)))))..(.---(....--------------....).)..)))((((........))))....))))))....------ ( -32.00, z-score = -2.03, R) >droYak2.chr3L 7381338 97 - 24197627 CGCUGAGUUAUCCAUCUAGCUUCGAGCUGUGACGUCACAGCUU---GUCAUAAGG---AAA----------------UAUAUACAGGGCGGUCAGAAAAGAGGCCAAAAUCGGCAAUGU------ .(((((....(((...((....(((((((((....))))))))---)...)).))---)..----------------............((((........))))....))))).....------ ( -28.30, z-score = -1.94, R) >droSec1.super_2 6729182 113 - 7591821 AGCUGAGCUAUCCAUCUAGUUGCGAGCUGUGACGUCACAGUUU---GUCAAAGGG---AAAUACAUACACACAUAUUUAUAUGCAGGGCGGUCAGAAAAGAGGCCAAAAUCGGCGAUGU------ .((((((((.....(((..((((((((((((....))))))))---).)))..))---)............((((....))))...)))((((........))))....))))).....------ ( -33.90, z-score = -2.27, R) >droSim1.chr3L 6274413 113 - 22553184 AGCUGAGCUAUCCAUCUAGUUGCGAGCUGUGACGUCACCGUUU---GUCAAAGGG---AAAUAAAUACAUACAUAUAUAUAUGCAGGGCGGUCAGAAAAGAGGCCAAAAUCGGCGAUGU------ .((((((((.....(((..((((((((.(((....))).))))---).)))..))---)............((((....))))...)))((((........))))....))))).....------ ( -29.30, z-score = -0.89, R) >consensus AGCUGAGCUAUCCAUCUAGCUGCGAGCUGUGACGUCACAGUUU___GUCAAAGGG___AAAUA_AUACA_ACAUAU_UAUAUGCAGGGCGGUCAGAAAAGAGGCCAAAAUCGGCGAUGU______ .(((((((((......))))...((((.(((....))).))))..............................................((((........))))....)))))........... (-11.95 = -11.93 + -0.02)
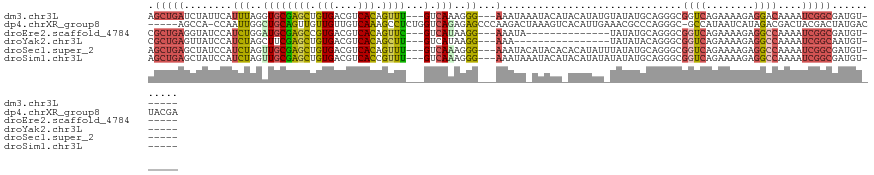
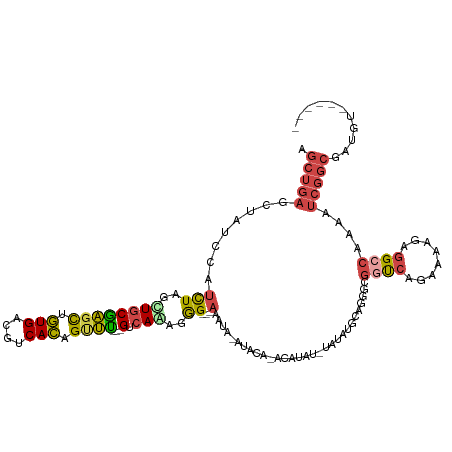

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:03 2011