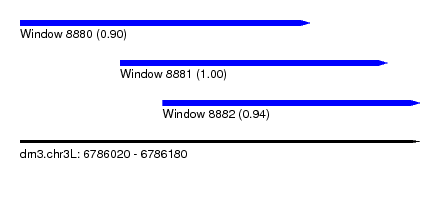
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,786,020 – 6,786,180 |
| Length | 160 |
| Max. P | 0.998400 |

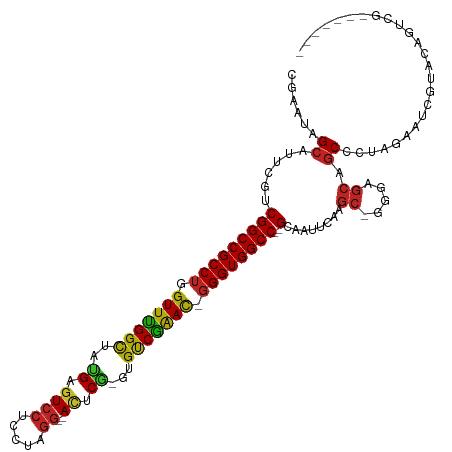
| Location | 6,786,020 – 6,786,136 |
|---|---|
| Length | 116 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.96 |
| Shannon entropy | 0.29175 |
| G+C content | 0.62239 |
| Mean single sequence MFE | -52.11 |
| Consensus MFE | -33.24 |
| Energy contribution | -32.65 |
| Covariance contribution | -0.59 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6786020 116 + 24543557 GGCACUUUAACAGCUUUGGCGGGGCGGGCGGAGCGGGUGGCGAAUAGCAUUCGUCGGCCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--AUUCG-GCGUCGAAU-GGGUGGCCGCAUUUC .((......((.((((((.(......).))))))..))(((((((...)))))))(((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))))..... ( -51.80, z-score = -1.87, R) >droSim1.chr3L 6265549 116 + 22553184 GGCACUUCAACAGCUUUGGCGGGGCGGGCGGAGCGGGUGGCGAAUAGCAUUCGUCGGCCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--AUUCG-GCGUCGAAU-GGGUGGCCGCAUUUC .((......((.((((((.(......).))))))..))(((((((...)))))))(((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))))..... ( -51.80, z-score = -1.71, R) >droYak2.chr3L 7372063 116 + 24197627 GGCACUUCAACAGCUUUGGCGGGGCGGGCGGUGCGGGUGGGGAAUAGCAUUCGUCGGCCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--AUUCG-GCGUCGAAC-GGGUGGCCGCAAUUC .(((((.(....((((.....))))..).)))))(((((........)))))(.((((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))))..... ( -52.50, z-score = -1.94, R) >droEre2.scaffold_4784 21188618 116 - 25762168 GGCACUUCAAUAGCUUUGGCGGGGCGGGCGGAGCGGGUGGGGAAUAGCAUUCGUCGGCCGCCUGGUUUGGCUAUGAGUCCUCCUAGG--ACUCG-GCGUCGAAC-GGGUGGCCGCAAUUC .((.(((((...((((((.(......).))))))...))))).............(((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))))..... ( -53.60, z-score = -2.38, R) >droAna3.scaffold_13337 3169059 116 - 23293914 GUCACUUCAACAGCUUCGGCGGGGCGGGCGGAGCGGGCGGCGAAUAGCAUUCGUCGGCCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--ACUCG-GUGUCGAAC-GGGUGGCCGCAGUUC ........(((.((((((.(......).))))))..(((((((((...)))))))(((((((((.((((((..(((((((.....))--)))))-..)))))))-)))))))))).))). ( -59.20, z-score = -3.19, R) >dp4.chrXR_group8 4075620 116 - 9212921 GUCACUUCAAUAGCUUUGGCGGUGCGGGCGGCGCCGGUGGGGAAUAGCAUUCGUCGGCCGCCUGGUUUGGCUACGAGUCCUCCUAGG--AUUCG-GUGUCGAAC-GGGUGGCCGCAUUUC .............((..(.((((((.....)))))).)..))..........(.((((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))))..... ( -53.80, z-score = -2.67, R) >droPer1.super_38 554256 116 + 801819 GUCACUUCAAUAGCUUUGGCGGUGCGGGCGGCGCCGGUGGGGAAUAGCAUUCGUCGGCCGCCUGGUUUGGCUACGAGUCCUCCUAGG--AUUCG-GUGUCGAAC-GGGUGGCCGCAUUUC .............((..(.((((((.....)))))).)..))..........(.((((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))))..... ( -53.80, z-score = -2.67, R) >droWil1.scaffold_180916 2319502 113 - 2700594 GUCAUUUCAAUAGCUUUGGCGGCGCUGGGGGAGCGGGCGGCGAAUAGCAUACAUCGGCCGCCUGGUUUGGCUAUGAAUCCUCUUAAACAACUCAUACGUCAAAUUGGGUGGCC------- ............(((....((.((((.(.....).)))).))...))).......(((((((..((((((((((((...............))))).)))))))..)))))))------- ( -44.06, z-score = -2.82, R) >droVir3.scaffold_13049 7125577 113 + 25233164 GUCAUUUCAAUAGCUUUGGCGGCG---GCGGUGCGGGCGGUGAAUAGCAUUCGUCGGCCGCCUGGUUUGGUUACGAGUCCUCCUAGG--ACACA-AUGCCAGAC-GGGUGGCCGGAAUGC .....((((...((((..(((.(.---..).)))))))..))))..((((((..((((((((((.((((((...(.((((.....))--)).).-..)))))))-))))))))))))))) ( -47.90, z-score = -1.89, R) >droMoj3.scaffold_6680 5401661 113 + 24764193 GUCACUUCAAUAGCUUUGGCGGCG---GCGGUUCGGGUGGUGAAUAGCAUUCGUCGGCCGCCUGGUUCGGCUACGAGUCCUCCUAGG--ACACG-CUGUCGGAC-GGGUGGCCGCAAUGC .(((((..(((.(((........)---)).)))..)))))......(((((.(.((((((((((.((((((..((.((((.....))--)).))-..)))))))-))))))))))))))) ( -50.40, z-score = -2.02, R) >droGri2.scaffold_15110 18841777 113 - 24565398 GUCAUUUCAAUAGCUUUGGGGGCG---GCGGUGCGGGUGGUGAAUAGCAUUCGUCGGCCGCCUGGUUUGGCUAUGAGUCCUCCUAGG--ACCCA-CUGCCGGAC-CGGUGGCCGCAUUGC (((...((((.....)))).))).---((((((((((((........)))))...(((((((.((((((((..((.((((.....))--)).))-..)))))))-))))))))))))))) ( -54.40, z-score = -2.68, R) >consensus GUCACUUCAAUAGCUUUGGCGGGGCGGGCGGAGCGGGUGGCGAAUAGCAUUCGUCGGCCGCCUGGUUUGGCUAUGAGUCCUCCUAGG__ACUCG_GUGUCGAAC_GGGUGGCCGCAUUUC ............((...(((((.....((....((.....))....))..)))))((((((((.(((((((..((.((((.....))..)).))...))))))).))))))))))..... (-33.24 = -32.65 + -0.59)

| Location | 6,786,060 – 6,786,167 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 79.27 |
| Shannon entropy | 0.41134 |
| G+C content | 0.58817 |
| Mean single sequence MFE | -46.38 |
| Consensus MFE | -26.35 |
| Energy contribution | -25.30 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.998400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6786060 107 + 24543557 CGAAUAGCAUUCGUCGGCCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--AUUCG-GCGUCGAAU-GGGUGGCC--GCAUUUCAAGC-GGGAGCAGCCCUAGAAUCGUACAGUCG------- (((...(((((((.((((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))--)).........-(((.....)))..)))).))....)))------- ( -44.20, z-score = -2.29, R) >droSim1.chr3L 6265589 107 + 22553184 CGAAUAGCAUUCGUCGGCCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--AUUCG-GCGUCGAAU-GGGUGGCC--GCAUUUCAAGC-GGGAGCAGCCCUAGAAUCGUACAGUCG------- (((...(((((((.((((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))--)).........-(((.....)))..)))).))....)))------- ( -44.20, z-score = -2.29, R) >droYak2.chr3L 7372103 107 + 24197627 GGAAUAGCAUUCGUCGGCCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--AUUCG-GCGUCGAAC-GGGUGGCC--GCAAUUCAAGC-GGGAGCAGCCCUAGAAUCGUACAGUCG------- .((...(((((((.((((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))--)).........-(((.....)))..)))).))....)).------- ( -46.00, z-score = -2.60, R) >droEre2.scaffold_4784 21188658 107 - 25762168 GGAAUAGCAUUCGUCGGCCGCCUGGUUUGGCUAUGAGUCCUCCUAGG--ACUCG-GCGUCGAAC-GGGUGGCC--GCAAUUCAAGC-GGGAGCAGCCCUAGAAUCGUACAGUCG------- .((...(((((((.((((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))--)).........-(((.....)))..)))).))....)).------- ( -46.20, z-score = -2.54, R) >droAna3.scaffold_13337 3169099 107 - 23293914 CGAAUAGCAUUCGUCGGCCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--ACUCG-GUGUCGAAC-GGGUGGCC--GCAGUUCAAGC-GGGAGCAGCCCUAGAGCCGAACAGUCG------- (((.........(.((((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))--)).((((..((-....)).((......)).))))..)))------- ( -49.80, z-score = -2.99, R) >dp4.chrXR_group8 4075660 96 - 9212921 GGAAUAGCAUUCGUCGGCCGCCUGGUUUGGCUACGAGUCCUCCUAGG--AUUCG-GUGUCGAAC-GGGUGGCC--GCAUUUCAAGC-GUGAGCCGC-----AGCCGUA------------- ((....((....(.((((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))--)).......((-....)).))-----..))...------------- ( -46.70, z-score = -3.64, R) >droPer1.super_38 554296 96 + 801819 GGAAUAGCAUUCGUCGGCCGCCUGGUUUGGCUACGAGUCCUCCUAGG--AUUCG-GUGUCGAAC-GGGUGGCC--GCAUUUCAAGC-GUGAGCCGC-----AGCCGUA------------- ((....((....(.((((((((((.((((((..(((((((.....))--)))))-..)))))))-))))))))--)).......((-....)).))-----..))...------------- ( -46.70, z-score = -3.64, R) >droWil1.scaffold_180916 2319542 104 - 2700594 CGAAUAGCAUACAUCGGCCGCCUGGUUUGGCUAUGAAUCCUCUUAAACAACUCAUACGUCAAAUUGGGUGGCCCAGGAACGCAAUCCGACAACCACUUUAGAUA----------------- ............((((((((((..((((((((((((...............))))).)))))))..)))))))..(((......))).............))).----------------- ( -33.66, z-score = -3.66, R) >droVir3.scaffold_13049 7125614 99 + 25233164 UGAAUAGCAUUCGUCGGCCGCCUGGUUUGGUUACGAGUCCUCCUAGG--ACACA-AUGCCAGAC-GGGUGGCC--GGAAUGCAAGC-GCGAGCAGCCCUAAAAGCG--------------- ......((((((..((((((((((.((((((...(.((((.....))--)).).-..)))))))-))))))))--)))))))..((-....)).((.......)).--------------- ( -46.70, z-score = -4.25, R) >droMoj3.scaffold_6680 5401698 107 + 24764193 UGAAUAGCAUUCGUCGGCCGCCUGGUUCGGCUACGAGUCCUCCUAGG--ACACG-CUGUCGGAC-GGGUGGCC--GCAAUGCAAGC-GCGAGCAGCCCUGAAAACCAGAUGCAG------- ......(((((.(.((((((((((.((((((..((.((((.....))--)).))-..)))))))-))))))))--)))))))..((-....)).((.(((.....)))..))..------- ( -51.30, z-score = -3.77, R) >droGri2.scaffold_15110 18841814 114 - 24565398 UGAAUAGCAUUCGUCGGCCGCCUGGUUUGGCUAUGAGUCCUCCUAGG--ACCCA-CUGCCGGAC-CGGUGGCC--GCAUUGCAAGC-GUGAGCAGCCCUAGAUUCGUAUCCUCGAAUCCAG ......(((...(.((((((((.((((((((..((.((((.....))--)).))-..)))))))-))))))))--))..)))..((-....))....((.((((((......)))))).)) ( -54.70, z-score = -5.41, R) >consensus CGAAUAGCAUUCGUCGGCCGCCUGGUUUGGCUAUGAGUCCUCCUAGG__ACUCG_GUGUCGAAC_GGGUGGCC__GCAAUUCAAGC_GGGAGCAGCCCUAGAAUCGUACAGUCG_______ ......((.......((((((((.(((((((..((.((((.....))..)).))...))))))).))))))))..((.......)).....))............................ (-26.35 = -25.30 + -1.05)
| Location | 6,786,077 – 6,786,180 |
|---|---|
| Length | 103 |
| Sequences | 11 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 70.66 |
| Shannon entropy | 0.55663 |
| G+C content | 0.58581 |
| Mean single sequence MFE | -35.83 |
| Consensus MFE | -19.56 |
| Energy contribution | -18.48 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6786077 103 + 24543557 CCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--AUUCG-GCGUCGAAU-GGGUGGCC--GCAUUUCAAGC-GGGAGCAGCCCUAGAAUCGUACAGUCGAACUUCUCUAACG----------- ......(((((((((.(((((((.....))--)))))-(((......-((((..((--((.......))-)).....))))......)))..))))))))).........----------- ( -34.70, z-score = -0.54, R) >droSim1.chr3L 6265606 103 + 22553184 CCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--AUUCG-GCGUCGAAU-GGGUGGCC--GCAUUUCAAGC-GGGAGCAGCCCUAGAAUCGUACAGUCGAACUUCUCUAACG----------- ......(((((((((.(((((((.....))--)))))-(((......-((((..((--((.......))-)).....))))......)))..))))))))).........----------- ( -34.70, z-score = -0.54, R) >droYak2.chr3L 7372120 103 + 24197627 CCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--AUUCG-GCGUCGAAC-GGGUGGCC--GCAAUUCAAGC-GGGAGCAGCCCUAGAAUCGUACAGUCGAACUUCUCUAACG----------- (((((((.((((((..(((((((.....))--)))))-..)))))))-))))))((--((.......))-))..........(((((((......)))..))))......----------- ( -36.80, z-score = -1.07, R) >droEre2.scaffold_4784 21188675 103 - 25762168 CCGCCUGGUUUGGCUAUGAGUCCUCCUAGG--ACUCG-GCGUCGAAC-GGGUGGCC--GCAAUUCAAGC-GGGAGCAGCCCUAGAAUCGUACAGUCGAACUUCUCUAACG----------- (((((((.((((((..(((((((.....))--)))))-..)))))))-))))))((--((.......))-))..........(((((((......)))..))))......----------- ( -37.00, z-score = -1.09, R) >droAna3.scaffold_13337 3169116 103 - 23293914 CCGCCUGGUUCGGCUAUGAGUCCUCCUAGG--ACUCG-GUGUCGAAC-GGGUGGCC--GCAGUUCAAGC-GGGAGCAGCCCUAGAGCCGAACAGUCGAGCUUCUCCUAUG----------- (((((((.((((((..(((((((.....))--)))))-..)))))))-)))))).(--((.......))-)((((........(((((((....))).))))))))....----------- ( -43.20, z-score = -1.50, R) >dp4.chrXR_group8 4075677 79 - 9212921 CCGCCUGGUUUGGCUACGAGUCCUCCUAGG--AUUCG-GUGUCGAAC-GGGUGGCC--GCAUUUCAAGC-GUGAGCCGC-----AGCCGUA------------------------------ ..(((((.((((((..(((((((.....))--)))))-..)))))))-))))(((.--((..((((...-.))))..))-----.)))...------------------------------ ( -33.00, z-score = -1.63, R) >droPer1.super_38 554313 79 + 801819 CCGCCUGGUUUGGCUACGAGUCCUCCUAGG--AUUCG-GUGUCGAAC-GGGUGGCC--GCAUUUCAAGC-GUGAGCCGC-----AGCCGUA------------------------------ ..(((((.((((((..(((((((.....))--)))))-..)))))))-))))(((.--((..((((...-.))))..))-----.)))...------------------------------ ( -33.00, z-score = -1.63, R) >droWil1.scaffold_180916 2319559 96 - 2700594 CCGCCUGGUUUGGCUAUGAAUCCUCUUAAACAACUCAUACGUCAAAUUGGGUGGCCCAGGAACGCAAUCCGACAACCACUUUAGAUAAGAUUUUUU------------------------- (((((..((((((((((((...............))))).)))))))..)))))....(((......)))..........................------------------------- ( -24.96, z-score = -2.03, R) >droVir3.scaffold_13049 7125631 82 + 25233164 CCGCCUGGUUUGGUUACGAGUCCUCCUAGG--ACACA-AUGCCAGAC-GGGUGGCC--GGAAUGCAAGC-GCGAGCAGCCCUAAAAGCG-------------------------------- (((((((.((((((...(.((((.....))--)).).-..)))))))-))))))..--((..(((....-....)))..))........-------------------------------- ( -29.80, z-score = -1.08, R) >droMoj3.scaffold_6680 5401715 94 + 24764193 CCGCCUGGUUCGGCUACGAGUCCUCCUAGG--ACACG-CUGUCGGAC-GGGUGGCC--GCAAUGCAAGC-GCGAGCAGCCCUGAAAACCAGAUGCAGAUCU-------------------- (((((((.((((((..((.((((.....))--)).))-..)))))))-))))))..--....((((.((-....))....(((.....))).)))).....-------------------- ( -37.80, z-score = -1.47, R) >droGri2.scaffold_15110 18841831 114 - 24565398 CCGCCUGGUUUGGCUAUGAGUCCUCCUAGG--ACCCA-CUGCCGGAC-CGGUGGCC--GCAUUGCAAGC-GUGAGCAGCCCUAGAUUCGUAUCCUCGAAUCCAGGACUUGGCUCUCCAACA (((((.((((((((..((.((((.....))--)).))-..)))))))-)))))).(--((.......))-).((((((.(((.((((((......)))))).))).))..))))....... ( -49.20, z-score = -3.54, R) >consensus CCGCCUGGUUUGGCUAUGAGUCCUCCUAGG__ACUCG_GUGUCGAAC_GGGUGGCC__GCAAUUCAAGC_GGGAGCAGCCCUAGAAUCGUACAGUCGAACU________G___________ ((((((.(((((((..((.((((.....))..)).))...))))))).))))))................................................................... (-19.56 = -18.48 + -1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:08:02 2011