| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,753,474 – 6,753,575 |
| Length | 101 |
| Max. P | 0.623626 |

| Location | 6,753,474 – 6,753,575 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.23 |
| Shannon entropy | 0.38929 |
| G+C content | 0.60764 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -28.52 |
| Energy contribution | -28.36 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.623626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
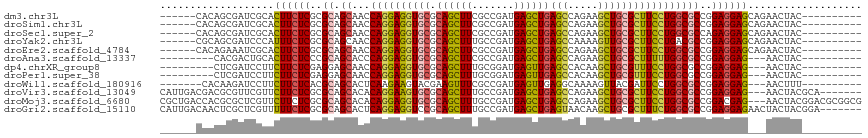
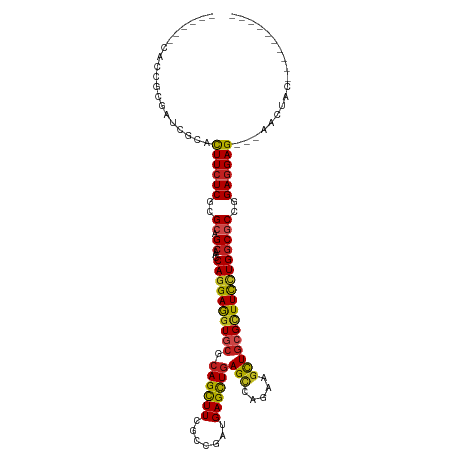
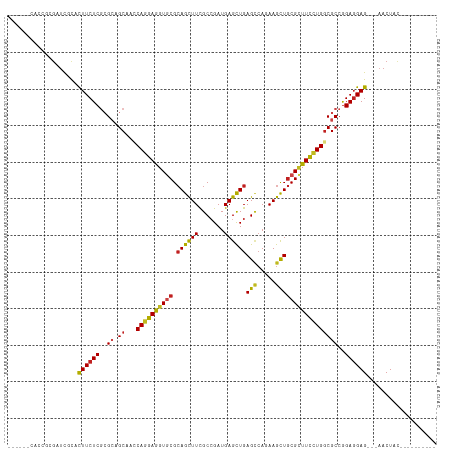
>dm3.chr3L 6753474 101 - 24543557 ------CACAGCGAUCGCACUUCUCGCGCAGCAACCAGGAGGUGCGCAGCUUCGCCGAUGAGCUGAGCCAGAAGCUGCGCUUCCUGGCGCCGGAGGAGCAGAACUAC---------- ------..........((.(((((.((((......((((((..((((((((((((((......)).))..)))))))))))))))))))).))))).))........---------- ( -41.90, z-score = -0.79, R) >droSim1.chr3L 6234963 101 - 22553184 ------CACAGCGAUCGCACUUCUCGCGCAGCAACCAGGAGGUGCGCAGCUUCGCCGAUGAGCUGAGCCAGAAGCUGCGCUUCCUGGCGCCGGAGGAGCAGAACUAC---------- ------..........((.(((((.((((......((((((..((((((((((((((......)).))..)))))))))))))))))))).))))).))........---------- ( -41.90, z-score = -0.79, R) >droSec1.super_2 6690201 101 - 7591821 ------CACAGCGAUCGCACUUCUCGCGCAGCAACCAGGAGGUGCGCAGCUUCGCCGAUGAGCUGAGCCAGAAGCUGCGCUUCCUGGCGCCAGAGGAGCAGAACUAC---------- ------..........((.(((((.((((......((((((..((((((((((((((......)).))..)))))))))))))))))))).))))).))........---------- ( -41.90, z-score = -1.10, R) >droYak2.chr3L 7338828 101 - 24197627 ------CGCAGCGAUCCCAUUUCUCGCGCAGCAACCAGGAGGUGCGCAGCUUUGCCGAUGAGCUGAGCCAAAAGUUGCGCUUCCUGACGCCGGAGGAGCAGAACUAC---------- ------.((.((((.........)))))).(((((.....(((...((((((.......)))))).)))....)))))((((((((....))).)))))........---------- ( -33.40, z-score = 0.22, R) >droEre2.scaffold_4784 21153711 101 + 25762168 ------CACAGAAAUCGCACUUCUCGCGCAGCAACCAGGAGGUGCGCAGCUUCGCCGAUGAGCUGAGCCAGAAGCUGCGCUUCCUGGCGCCGGAGGAGCAGAACUAC---------- ------..........((.(((((.((((......((((((..((((((((((((((......)).))..)))))))))))))))))))).))))).))........---------- ( -41.90, z-score = -1.43, R) >droAna3.scaffold_13337 3135810 95 + 23293914 ---------CACGACUGCACUUCUCCCGCAGCACCCAGGAGGUGCGCAGCUUCGCCGAUGAGCUGAGCCAGAAGCUGCGCUUUUUGGCGCCGGAGGAG---AACUAC---------- ---------..........(((((((.((.(((((.....)))))((((((((((((......)).))..))))))))(((....))))).)))))))---......---------- ( -40.30, z-score = -2.06, R) >dp4.chrXR_group8 4036613 95 + 9212921 ---------CUCGAUCCUUCUUCUCGAGGAGCAACCAGGAGGUGCGCAGCUUUGCGGAUGAGUUGAGCCACAAGCUGCGUUUCCUGGCGCCGGAGGAG---AACUAC---------- ---------........((((((((..((.....(((((((..(((((((((.(.((..........)).)))))))))))))))))..)).))))))---))....---------- ( -38.40, z-score = -1.60, R) >droPer1.super_38 515618 95 - 801819 ---------CUCGAUCCUUCUUCUCGAGGAGCAACCAGGAGGUGCGCAGCUUUGCGGAUGAGUUGAGCCACAAGCUGCGUUUCCUGGCGCCGGAGGAG---AACUAC---------- ---------........((((((((..((.....(((((((..(((((((((.(.((..........)).)))))))))))))))))..)).))))))---))....---------- ( -38.40, z-score = -1.60, R) >droWil1.scaffold_180916 2268772 97 + 2700594 -------CACAAGAUCCUUCUUCUCACGCAGCACUCAAGAAGUACGAAGUUUCGCCGAUGAGUUGAGGCAAAAGUUACGAUUCCUGGCGCCGGAGGAG---AACUUU---------- -------..........((((((((.((..((......((((.......))))((((..((((((..((....))..)))))).))))))))))))))---))....---------- ( -25.30, z-score = -0.29, R) >droVir3.scaffold_13049 7087505 107 - 25233164 CAUUGACGACGCGUUCGUUCUUCUCGCGCAGCACACAGGAAGUGCGCAGCUUUGCCGAUGAGCUGAGCCAGAAGCUGCGCUUCCUGGCGCCGGAGGAG---AACUACGCA------- ..........((((..(((((((((.((..((.(.((((((((((.((((((.......))))))(((.....)))))))))))))).))))))))))---))).)))).------- ( -50.80, z-score = -3.08, R) >droMoj3.scaffold_6680 5363978 114 - 24764193 CGCUGACCACGCGCUCGUUCUUCUCGCGCAGCACACAGGAGGUGCGCAGCUUUGCCGAUGAGCUGAGCCAGAAGCUGCGCUUCCUGGCGCCGGACGAG---AACUACGGACGCGGCG .........((((..(((..(((((((.(.((.(.((((((((((.((((((.......))))))(((.....)))))))))))))).)).)).))))---))..)))..))))... ( -52.40, z-score = -1.47, R) >droGri2.scaffold_15110 18806911 110 + 24565398 CAUUGACAACUCGCUCGUUUUUCUCGCGCAGCACUCAGGAGGUCCGCAGCUUUGCCGAUGAGCUGAGUAACAAGCUGCGCUUUCUGGCGCCGGAGGAGAACUACUACGGA------- ..........(((...((..((((((((((((((((((...(((.((......)).)))...)))))).....))))))).(((((....))))))))))..))..))).------- ( -37.00, z-score = -0.15, R) >consensus ______CACCGCGAUCGCACUUCUCGCGCAGCAACCAGGAGGUGCGCAGCUUCGCCGAUGAGCUGAGCCAGAAGCUGCGCUUCCUGGCGCCGGAGGAG___AACUAC__________ ...................((((((..((.((...((((((((((.((((((.......))))))(((.....)))))))))))))))))..))))))................... (-28.52 = -28.36 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:53 2011