| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,749,831 – 6,749,933 |
| Length | 102 |
| Max. P | 0.999967 |

| Location | 6,749,831 – 6,749,933 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 63.53 |
| Shannon entropy | 0.72243 |
| G+C content | 0.55638 |
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -15.19 |
| Energy contribution | -15.14 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
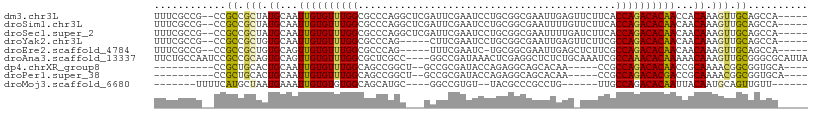
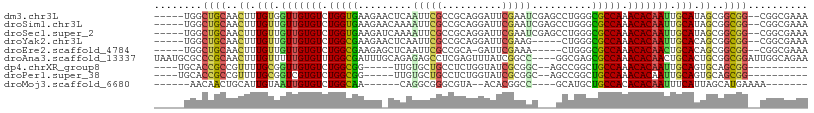
>dm3.chr3L 6749831 102 + 24543557 UUUCGCCG--CCGCCGCUAUGCAAUUGUGUUUGGCGCCCAGGCUCGAUUCGAAUCCUGCGGCGAAUUGAGUUCUUCACCAGACACAACCACAAAGUUGCAGCCA----- .......(--(.((.(((.((...((((((((((((((((((.(((...)))..)))).))))...((((...))))))))))))))...)).))).)).))..----- ( -34.40, z-score = -2.04, R) >droSim1.chr3L 6231260 102 + 22553184 UUUCGCCG--CCGCCGCUAUGCAAUUGUGUUUGGCGCCCAGGCUCGAUUCGAAUCCUGCGGCGAAUUUUGUUCUUCACCAGACACAACAACAAAGUUGCAGCCA----- .......(--(.((.(((.((...((((((((((.(((((((.(((...)))..)))).)))((((...))))....))))))))))...)).))).)).))..----- ( -33.60, z-score = -2.01, R) >droSec1.super_2 6686479 102 + 7591821 UUUCGCCG--CCGCCGCUAUGCAAUUGUGUUUGGCGCCCAGGCUCGAUUCGAAUCCUGCGGCGAAUUUUGAUCUUCACCAGACACAACAACAAAGUUGCAGCCA----- .......(--(.((.(((.((...((((((((((((((((((.(((...)))..)))).)))).....((.....))))))))))))...)).))).)).))..----- ( -34.10, z-score = -2.21, R) >droYak2.chr3L 7335108 97 + 24197627 UUUCGCCG--CCGCCGCUGUGCAAUUGUGUUUGGCGCCCAG-----CUUCGAAUCCUGCGGCGAAUUGAGUUCUUCGCCAGACACAACAACAAAGUUGCAGCCA----- .......(--(.((.(((.((...(((((((((((((((((-----.........))).)))((((...))))...)))))))))))...)).))).)).))..----- ( -33.00, z-score = -1.42, R) >droEre2.scaffold_4784 21149964 96 - 25762168 UUUCGCCG--CCGCCGCUGUGCAGUUGUGUUUGGCGCCCAG-----UUUCGAAUC-UGCGGCGAAUUGAGCUCUUCGCCAGACACAACAACAAAGUUGCAGCCA----- .......(--(.((.(((.((..((((((((((((((((((-----........)-)).)))(((........)))))))))))))))..)).))).)).))..----- ( -38.40, z-score = -2.74, R) >droAna3.scaffold_13337 3131575 105 - 23293914 UUCUGCCAAUCCGCCGCAGUGCAGUUGUGUUUGGCGCUCGCC----GGCCGAUAAACUCGAGGCUCUCUGCAAAUCGCCAAACACAAAAACAAAGUUGCGGGCGCAUUA ...(((...(((((.((..((...((((((((((((...((.----((((...........))))....))....))))))))))))...))..)).))))).)))... ( -37.60, z-score = -1.78, R) >dp4.chrXR_group8 4032937 88 - 9212921 ----------CCGCUGCACUGCAAUUGUGUUUGGCAGCCGGCU--GCCGCGAUACCAGAGGCAGCACAA-----CCGCCAGACACAACCGCAAAACGGCGGUGCA---- ----------((((((...(((..(((((((((((.(...(((--(((.(.......).))))))....-----).)))))))))))..)))...))))))....---- ( -42.80, z-score = -3.59, R) >droPer1.super_38 511862 88 + 801819 ----------CCGCUGCACUGCAAUUGUGUUUGGCAGCCGGCU--GCCGCGAUACCAGAGGCAGCACAA-----CCGCCAGACACGACCGCAAAACGGCGGUGCA---- ----------((((((...(((..(((((((((((.(...(((--(((.(.......).))))))....-----).)))))))))))..)))...))))))....---- ( -42.50, z-score = -3.35, R) >droMoj3.scaffold_6680 5359766 84 + 24764193 -------UUUUCAUGCUAAUGAAAUUGUGUGUGGCAGCAUGC----GGCCGUGU--UACGCCCGCCUG------UUGCCAGACACAAUUACAAUGCAGUUGUU------ -------......(((...((.((((((((.((((((((.((----((.((...--..)).)))).))------)))))).)))))))).))..)))......------ ( -39.10, z-score = -5.30, R) >consensus UUUCGCCG__CCGCCGCUAUGCAAUUGUGUUUGGCGCCCAGC___GAUUCGAAUCCUGCGGCGAAUUGAG_UCUUCGCCAGACACAACAACAAAGUUGCAGCCA_____ ............((.(((.((...((((((((((...........................................))))))))))...)).))).)).......... (-15.19 = -15.14 + -0.04)
| Location | 6,749,831 – 6,749,933 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 63.53 |
| Shannon entropy | 0.72243 |
| G+C content | 0.55638 |
| Mean single sequence MFE | -44.14 |
| Consensus MFE | -23.57 |
| Energy contribution | -23.49 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.58 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.36 |
| SVM RNA-class probability | 0.999967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
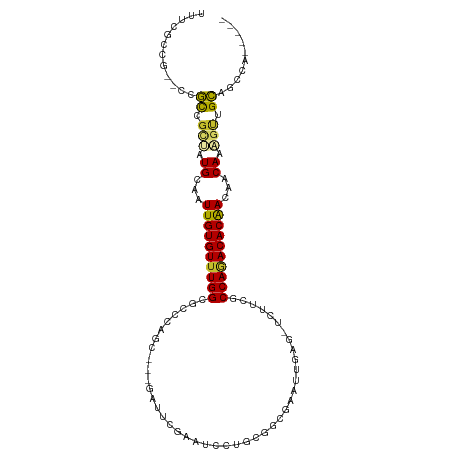
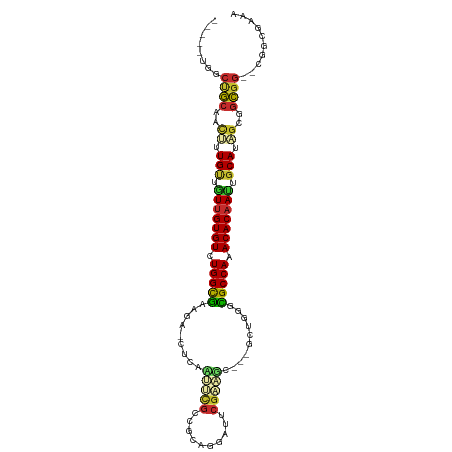
>dm3.chr3L 6749831 102 - 24543557 -----UGGCUGCAACUUUGUGGUUGUGUCUGGUGAAGAACUCAAUUCGCCGCAGGAUUCGAAUCGAGCCUGGGCGCCAAACACAAUUGCAUAGCGGCGG--CGGCGAAA -----..(((((..((.((..(((((((.(((((((........)))(((.((((.((((...))))))))))))))).)))))))..)).))..))))--)....... ( -46.90, z-score = -3.96, R) >droSim1.chr3L 6231260 102 - 22553184 -----UGGCUGCAACUUUGUUGUUGUGUCUGGUGAAGAACAAAAUUCGCCGCAGGAUUCGAAUCGAGCCUGGGCGCCAAACACAAUUGCAUAGCGGCGG--CGGCGAAA -----..(((((..((.(((.(((((((.(((((((........)))(((.((((.((((...))))))))))))))).))))))).))).))..))))--)....... ( -43.80, z-score = -3.39, R) >droSec1.super_2 6686479 102 - 7591821 -----UGGCUGCAACUUUGUUGUUGUGUCUGGUGAAGAUCAAAAUUCGCCGCAGGAUUCGAAUCGAGCCUGGGCGCCAAACACAAUUGCAUAGCGGCGG--CGGCGAAA -----..(((((..((.(((.(((((((.(((((((........)))(((.((((.((((...))))))))))))))).))))))).))).))..))))--)....... ( -43.80, z-score = -3.29, R) >droYak2.chr3L 7335108 97 - 24197627 -----UGGCUGCAACUUUGUUGUUGUGUCUGGCGAAGAACUCAAUUCGCCGCAGGAUUCGAAG-----CUGGGCGCCAAACACAAUUGCACAGCGGCGG--CGGCGAAA -----..(((((..((.(((.(((((((.(((((((........)))(((.(((.........-----)))))))))).))))))).))).))..))))--)....... ( -39.60, z-score = -2.05, R) >droEre2.scaffold_4784 21149964 96 + 25762168 -----UGGCUGCAACUUUGUUGUUGUGUCUGGCGAAGAGCUCAAUUCGCCGCA-GAUUCGAAA-----CUGGGCGCCAAACACAACUGCACAGCGGCGG--CGGCGAAA -----..(((((..((.(((.(((((((.(((((((........)))(((.((-(........-----)))))))))).))))))).))).))..))))--)....... ( -41.50, z-score = -2.74, R) >droAna3.scaffold_13337 3131575 105 + 23293914 UAAUGCGCCCGCAACUUUGUUUUUGUGUUUGGCGAUUUGCAGAGAGCCUCGAGUUUAUCGGCC----GGCGAGCGCCAAACACAACUGCACUGCGGCGGAUUGGCAGAA ((((.((((.(((....(((..((((((((((((.(((((...(.(((..((.....))))))----.)))))))))))))))))..))).))))))).))))...... ( -44.50, z-score = -2.93, R) >dp4.chrXR_group8 4032937 88 + 9212921 ----UGCACCGCCGUUUUGCGGUUGUGUCUGGCGG-----UUGUGCUGCCUCUGGUAUCGCGGC--AGCCGGCUGCCAAACACAAUUGCAGUGCAGCGG---------- ----....((((.((.((((((((((((.((((((-----(((.((((((.(.......).)))--)))))))))))).)))))))))))).)).))))---------- ( -53.20, z-score = -5.27, R) >droPer1.super_38 511862 88 - 801819 ----UGCACCGCCGUUUUGCGGUCGUGUCUGGCGG-----UUGUGCUGCCUCUGGUAUCGCGGC--AGCCGGCUGCCAAACACAAUUGCAGUGCAGCGG---------- ----....((((.((.(((((((.((((.((((((-----(((.((((((.(.......).)))--)))))))))))).)))).))))))).)).))))---------- ( -49.10, z-score = -3.98, R) >droMoj3.scaffold_6680 5359766 84 - 24764193 ------AACAACUGCAUUGUAAUUGUGUCUGGCAA------CAGGCGGGCGUA--ACACGGCC----GCAUGCUGCCACACACAAUUUCAUUAGCAUGAAAA------- ------......(((..((.((((((((.(((((.------((.((((.((..--...)).))----)).)).))))).)))))))).))...)))......------- ( -34.90, z-score = -4.47, R) >consensus _____UGGCUGCAACUUUGUUGUUGUGUCUGGCGAAGA_CUCAAUUCGCCGCAGGAUUCGAAGC___GCUGGGCGCCAAACACAAUUGCAUAGCGGCGG__CGGCGAAA ........((((..((.(((.(((((((.(((((.........(((((..........)))))..........))))).))))))).))).))..)))).......... (-23.57 = -23.49 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:52 2011