| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,749,546 – 6,749,606 |
| Length | 60 |
| Max. P | 0.526671 |

| Location | 6,749,546 – 6,749,606 |
|---|---|
| Length | 60 |
| Sequences | 9 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 67.84 |
| Shannon entropy | 0.65523 |
| G+C content | 0.35445 |
| Mean single sequence MFE | -9.21 |
| Consensus MFE | -5.88 |
| Energy contribution | -5.44 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.02 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.526671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
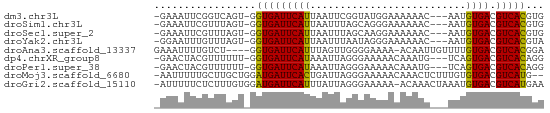
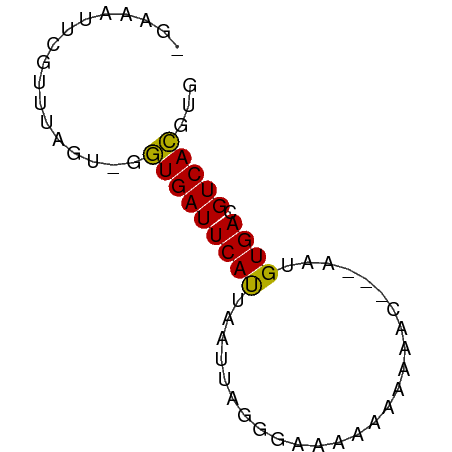
>dm3.chr3L 6749546 60 + 24543557 -GAAAUUCGGUCAGU-GGUGAUUCAUUAAUUCGGUAUGGAAAAAAC---AAUGUGACGUCACGUG -.....((((..(((-((....)))))...))))............---.(((((....))))). ( -7.90, z-score = 1.13, R) >droSim1.chr3L 6230974 60 + 22553184 -GAAAUUCGUUUAGU-GGUGAUUCAUUAAUUUAGCAGGGAAAAAAC---AAUGUGACGUCACGUG -.........(((((-((....))))))).................---.(((((....))))). ( -7.70, z-score = 0.60, R) >droSec1.super_2 6686193 60 + 7591821 -GAAAUUCGUUUAGU-GGUGAUUCAUUAAUUUAGCAAGGAAAAAAC---AAUGUGACGUCACGUG -.........(((((-((....))))))).................---.(((((....))))). ( -7.70, z-score = 0.51, R) >droYak2.chr3L 7334826 60 + 24197627 -GGAAUUUGUUUAGU-GGUGAUUCAUUAAUUUAAUAGGGAAAAAAC---AAUGUGACGUCACGUA -...(((...(((((-((....)))))))...)))...........---.(((((....))))). ( -7.90, z-score = 0.00, R) >droAna3.scaffold_13337 3131080 60 - 23293914 GAAAUUUUGUCU----GGUGAUUCAUUUAGUUGGGGAAAA-ACAAUUGUUUUGUGACGUCACGGA .........(((----(.((((((((.((((((.......-.))))))....)))).)))))))) ( -12.10, z-score = -0.70, R) >dp4.chrXR_group8 4032568 60 - 9212921 -GAACUACGUUUUUU-GGUGAUUCAUAAAUUAGGGAAAAACAAAUG---UCAGUGACGUCACAGG -.......(((((((-..(((((....)))))..))))))).....---...(((....)))... ( -9.30, z-score = -0.19, R) >droPer1.super_38 511493 60 + 801819 -GAACUACGUUUUUU-GGUGAUUCAUAAAUUAGGGAAAAACAAAUG---UCAGUGACGUCACAGG -.......(((((((-..(((((....)))))..))))))).....---...(((....)))... ( -9.30, z-score = -0.19, R) >droMoj3.scaffold_6680 19407427 62 - 24764193 -AAUUUUUGCUUGCUGGAUGAUUCACUGAUUAGGGAAAAACAAACUCUUUGUGUGACGUCAUG-- -................(((((((((.....((((.........))))....)))).))))).-- ( -9.30, z-score = -0.07, R) >droGri2.scaffold_15110 5765797 63 + 24565398 -AUUUUUCUCUUUGUGGAUGAUUCAUUUAUUAGGGAAAAA-ACAAACUAAAUGUGACGUCAUGAA -.(((((((((..(((((((...))))))).)))))))))-...........(((....)))... ( -11.70, z-score = -1.28, R) >consensus _GAAAUUCGUUUAGU_GGUGAUUCAUUAAUUAGGGAAAAAAAAAAC___AAUGUGACGUCACGUG .................(((((((((..........................)))).)))))... ( -5.88 = -5.44 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:50 2011