| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,713,841 – 6,713,944 |
| Length | 103 |
| Max. P | 0.969088 |

| Location | 6,713,841 – 6,713,944 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.01 |
| Shannon entropy | 0.49938 |
| G+C content | 0.43366 |
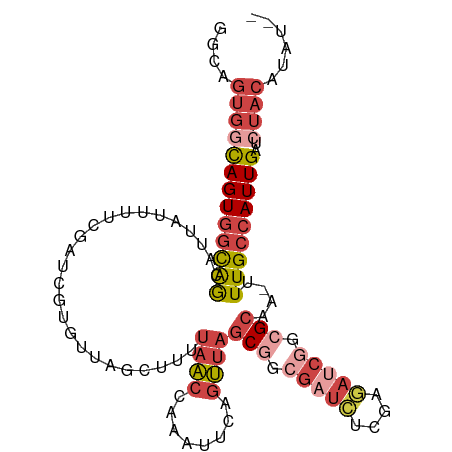
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -15.75 |
| Energy contribution | -18.03 |
| Covariance contribution | 2.28 |
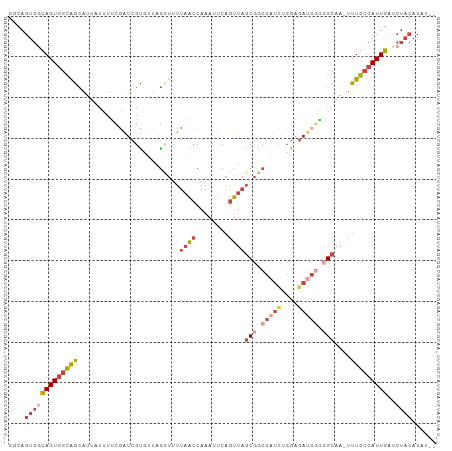
| Combinations/Pair | 1.24 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
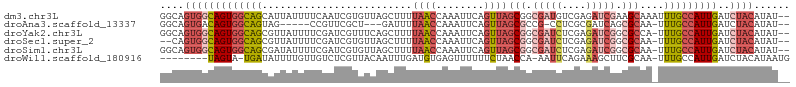
>dm3.chr3L 6713841 103 + 24543557 GGCAGUGGCAGUGGCAGCAUUAUUUUCAAUCGUGUUAGCUUUUAACCAAAUUCAGUUAGCGGCGAUGUCGAGAUCGAAGCAAAUUUGCCAUUGAUCUACAUAU-- ....((((((((((((((.((..((((.((((((((((((.............))))))).)))))...))))..)).)).....))))))))..))))....-- ( -27.72, z-score = -1.59, R) >droAna3.scaffold_13337 3094084 93 - 23293914 GGCAGUGACAGUGGCAGUAG-----CCGUUCGCU---GAUUUUAACCAAAUUCAGUUAGCGCCG-CCUCGCGAUCAGCGCAA-UUUGCCAUUGAUCUACAUAU-- ....(((((((((((((..(-----(((((.(((---((.(((....))).))))).))))..)-)..)(((.....)))..-..)))))))).)).))....-- ( -28.50, z-score = -1.83, R) >droYak2.chr3L 7298277 102 + 24197627 GGCAGUGGCAGUGGCAGCGUUAUUUUCGAUCGUUUCAGCUUUUAACCAAAUUCAGUUAGCGGCGAUCUCGAGAUCGGCGCCA-UUUGCCAUUGAUCUACAUAU-- (((((((((((((((.((...................(((.(((((........))))).)))((((....)))).))))))-.))))))))).)).......-- ( -32.60, z-score = -2.49, R) >droSec1.super_2 6650173 100 + 7591821 --CAGUGGCAGUGGCAGCGUUAUUUUCGAUCGUGUUAGCUUUUAACCAAAUUCAGUUAGCGGCGAUCUCGAGAUCGGCGCAA-UUUGCCAUUGAUCUACAUAU-- --..(((((((((((((((((..(((((((((((((((((.............))))))).))))..))))))..)))))..-..))))))))..))))....-- ( -35.52, z-score = -3.86, R) >droSim1.chr3L 6198745 102 + 22553184 GGCAGUGGCAGUGGCAGCGAUAUUUUCGAUCGUGUUAGCUUUUAACCAAAUUCAGUUAGCGGCGAUCUCGAGAUCGGCGCAA-UUUGCCAUUGAUCUACAUAU-- ....(((((((((((((((((.......))))).........................(((.(((((....))))).)))..-..))))))))..))))....-- ( -33.10, z-score = -2.31, R) >droWil1.scaffold_180916 2216001 94 - 2700594 --------UAGUA-UGAUAUUUUGUUGUCUCGUUACAAUUUGAUGUGAGUUUUUUCUAACCA-AAUUCAGAAAGCUUCGCAA-UUUGCCAUUGAUCUACAUAAUG --------..(((-.((((((..(((((......)))))..)))(((((((((((.......-.....))))))).))))..-..........))))))...... ( -12.60, z-score = 0.45, R) >consensus GGCAGUGGCAGUGGCAGCAUUAUUUUCGAUCGUGUUAGCUUUUAACCAAAUUCAGUUAGCGGCGAUCUCGAGAUCGGCGCAA_UUUGCCAUUGAUCUACAUAU__ ....(((((((((((((.........................((((........))))(((.(((((....))))).)))....)))))))))..))))...... (-15.75 = -18.03 + 2.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:48 2011