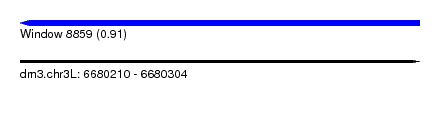
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,680,210 – 6,680,304 |
| Length | 94 |
| Max. P | 0.914708 |

| Location | 6,680,210 – 6,680,304 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.15 |
| Shannon entropy | 0.49351 |
| G+C content | 0.46890 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -13.53 |
| Energy contribution | -13.76 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
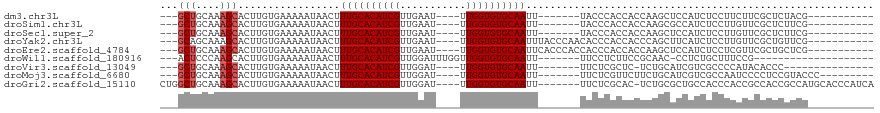
>dm3.chr3L 6680210 94 - 24543557 ---GCUGCAAAGCACUUGUGAAAAAUAACUUUGCACAUCGUUGAAU----UUGGUGUGCAAUU-------UACCCACCACCAAGCUCCAUCUCCUUCUUCGCUCUACG----------- ---(((....))).((((((..........((((((((((......----.))))))))))..-------.......)).))))........................----------- ( -18.11, z-score = -1.78, R) >droSim1.chr3L 6164919 94 - 22553184 ---GCUGCAAAGCACUUGUGAAAAAUAACUUUGCACAUCGUUGAAU----UUGGUGUGCAAUU-------UACCCACCACCAAGCGCCAUCUCCUUGUUCGCUCUUCG----------- ---(((....)))....(((((........((((((((((......----.))))))))))..-------..........((((.(......))))))))))......----------- ( -20.50, z-score = -1.44, R) >droSec1.super_2 6616766 94 - 7591821 ---GCUGCAAAGCACUUGUGAAAAAUAACUUUGCACAUCGUUGAAU----UUGGUGUGCAAUU-------UACCCACCACCAAGCUCCAUCUCCUUGUUCGCUCUUCG----------- ---(((....)))....(((((........((((((((((......----.))))))))))..-------..........((((.........)))))))))......----------- ( -20.20, z-score = -1.95, R) >droYak2.chr3L 7260383 101 - 24197627 ---GCAGCAAAGCACUUGUGAAAAAUAACUUUGCACAUCGUUGAAU----UUGGUGUGCAAUUUACCCAACACCCACCACCCAGCUUCAUCUCCUUGUUCGCUGUUCG----------- ---(((((.((((..((((.....))))..((((((((((......----.))))))))))......................))))((......))...)))))...----------- ( -21.00, z-score = -1.37, R) >droEre2.scaffold_4784 21076283 101 + 25762168 ---GCUGCAAAGCACUUGUGAAAAAUAACUUUGCACAUCGUUGAAU----UUGGUGUGCAAUUCACCCACCACCCACCACCAAGCUCCAUCUCCUCGUUCGCUGCUCG----------- ---(((....)))....(((((........((((((((((......----.)))))))))))))))................(((...............))).....----------- ( -18.46, z-score = -0.78, R) >droWil1.scaffold_180916 2127105 88 + 2700594 ---ACUCCCAAGCACUUGUGAAAAAUAACUUUGCACAUCGUUGGAUUUGGUUGGUGUGCAAUU-------UUCCUCUUCCGCAAC-CCUCUGCUUUCCG-------------------- ---......(((((.(((((..........((((((((((...........))))))))))..-------.........))))).-....)))))....-------------------- ( -17.70, z-score = -1.18, R) >droVir3.scaffold_13049 1004153 89 - 25233164 ---GCUGCAAAGCACUUGUGAAAAAUAACUUUGCACAUCGUUGGAU----UUGGUGUGCAAUU-------UUCUCGCUC-UCUGCAUCGUCGCCCCAUACACCC--------------- ---((.((...(((...((((.(((.....((((((((((......----.)))))))))).)-------)).))))..-..)))...)).))...........--------------- ( -21.10, z-score = -1.41, R) >droMoj3.scaffold_6680 24492579 96 + 24764193 ---GCUGCAAAGCACUUGUGAAAAAUAACUUUGCACAUCGUUGGAU----UUGGUGUGCAAUU-------UUCUCGUUCUUCUGCAUCGUCGCCAAUCCCCUCCGUACCC--------- ---(.(((((((.((....(((((......((((((((((......----.))))))))))))-------)))..)).))).)))).)......................--------- ( -19.10, z-score = -0.87, R) >droGri2.scaffold_15110 22394028 107 + 24565398 CUGGCUGCAAAGCACUUGUGAAAAAUAACUUUGCACAUCGUUGGAU----UUGGUGUGCAAUU-------UUCUCGCAC-UCUGCGCUGCCACCCACCGCCACCGCCAUGCACCCAUCA .(((.((((..((...(((((.(((.....((((((((((......----.)))))))))).)-------)).))))).-...(((.((.....)).)))....))..)))).)))... ( -30.70, z-score = -1.45, R) >consensus ___GCUGCAAAGCACUUGUGAAAAAUAACUUUGCACAUCGUUGAAU____UUGGUGUGCAAUU_______UACCCACCACCCAGCUCCAUCUCCUUGUUCGCUCUUCG___________ ...(((....))).................((((((((((...........)))))))))).......................................................... (-13.53 = -13.76 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:43 2011