
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,661,069 – 6,661,157 |
| Length | 88 |
| Max. P | 0.991591 |

| Location | 6,661,069 – 6,661,157 |
|---|---|
| Length | 88 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 76.20 |
| Shannon entropy | 0.43336 |
| G+C content | 0.61577 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -21.73 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
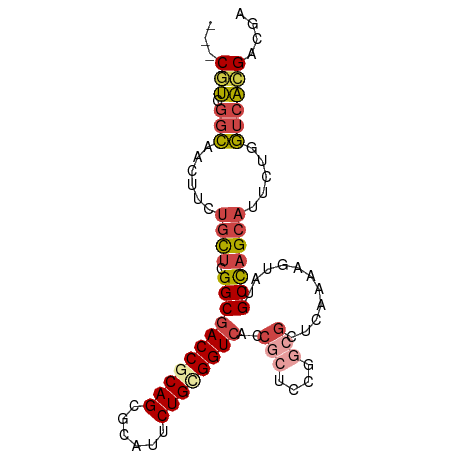
>dm3.chr3L 6661069 88 + 24543557 ---CGUCGGCAACUUCUGCUCGGCGACCGCAGCGCAUACUGCGGUCACCGCUCCGGCGCUCAAAAGUAUGCUAGCAUUCUGGUCACGCCGA ---.(((((((.....))).))))((((((((......)))))))).......(((((......((.(((....))).)).....))))). ( -35.10, z-score = -1.82, R) >droAna3.scaffold_13337 3022906 81 - 23293914 CUCCCGCACUGGAAUUUGUCUGGCGACCGCAGAGCAUUCUGUGGUCGCC-------CGCUGCCUGGCUCGCCAG---UCUGAUCCGGAGAC ((((((.(((((.........(((((((((((......)))))))))))-------.(((....)))...))))---).))....)))).. ( -36.10, z-score = -2.63, R) >droEre2.scaffold_4784 21057399 73 - 25762168 ---CGUCGGCAACUUCUGCUCGGCGACCACAGCGCAUUCUGCGGU---------------GAAAAGUAUGCCAGCAUUCUGGUCAGGACGA ---.(((((((.....))).))))((((.((.(((.....))).)---------------)...((.(((....))).))))))....... ( -23.50, z-score = -0.46, R) >droYak2.chr3L 7240631 88 + 24197627 ---CGUCGGCAACUUCUGCUCGGCGACCACAGCGCAUUCUGCGGUCACCGCUCCGGCGCUGAAAAGUAUGCCAGCAUUCUGGUCACGACAA ---.(((((((.....))).))))((((.((((((.....((((...))))....))))))...((.(((....))).))))))....... ( -34.10, z-score = -2.21, R) >droSec1.super_2 6597950 88 + 7591821 ---CGUCGGCAACUUCUGCUCGGCGACCGCAGCGCAUUCUGCGGUCACCGCUCCGGCGCUCAAAAGUAUGCUAGCAUUCUGGUCACGCCGA ---.(((((((.....))).))))((((((((......)))))))).......(((((......((.(((....))).)).....))))). ( -34.50, z-score = -1.67, R) >droSim1.chr3L 6143986 88 + 22553184 ---CGUCGGCAACUUCUGCUCGGCGACCGCAGCGCAUUCUGCGGUCACCGCUCCGGCGCUCAAAAGUAUGCUAGCAUUCUGGUCACGCCGA ---.(((((((.....))).))))((((((((......)))))))).......(((((......((.(((....))).)).....))))). ( -34.50, z-score = -1.67, R) >consensus ___CGUCGGCAACUUCUGCUCGGCGACCGCAGCGCAUUCUGCGGUCACCGCUCCGGCGCUCAAAAGUAUGCCAGCAUUCUGGUCACGACGA ...(((.(((......((((.(((((((((((......))))))))..(((....)))...........))))))).....)))))).... (-21.73 = -22.32 + 0.59)
| Location | 6,661,069 – 6,661,157 |
|---|---|
| Length | 88 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 76.20 |
| Shannon entropy | 0.43336 |
| G+C content | 0.61577 |
| Mean single sequence MFE | -36.25 |
| Consensus MFE | -24.42 |
| Energy contribution | -26.20 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6661069 88 - 24543557 UCGGCGUGACCAGAAUGCUAGCAUACUUUUGAGCGCCGGAGCGGUGACCGCAGUAUGCGCUGCGGUCGCCGAGCAGAAGUUGCCGACG--- ((((((........(((....)))(((((((..(((....))((((((((((((....)))))))))))))..)))))))))))))..--- ( -41.90, z-score = -3.22, R) >droAna3.scaffold_13337 3022906 81 + 23293914 GUCUCCGGAUCAGA---CUGGCGAGCCAGGCAGCG-------GGCGACCACAGAAUGCUCUGCGGUCGCCAGACAAAUUCCAGUGCGGGAG .((.(((.((..((---((((....))))......-------(((((((.((((....)))).)))))))........))..)).))))). ( -33.70, z-score = -1.64, R) >droEre2.scaffold_4784 21057399 73 + 25762168 UCGUCCUGACCAGAAUGCUGGCAUACUUUUC---------------ACCGCAGAAUGCGCUGUGGUCGCCGAGCAGAAGUUGCCGACG--- .((((((....))......((((.((((((.---------------(((((((......))))))).((...))))))))))))))))--- ( -21.80, z-score = -0.49, R) >droYak2.chr3L 7240631 88 - 24197627 UUGUCGUGACCAGAAUGCUGGCAUACUUUUCAGCGCCGGAGCGGUGACCGCAGAAUGCGCUGUGGUCGCCGAGCAGAAGUUGCCGACG--- ..((((((.((((....)))))))......((((....(..((((((((((((......))))))))))))..)....))))..))).--- ( -38.90, z-score = -2.75, R) >droSec1.super_2 6597950 88 - 7591821 UCGGCGUGACCAGAAUGCUAGCAUACUUUUGAGCGCCGGAGCGGUGACCGCAGAAUGCGCUGCGGUCGCCGAGCAGAAGUUGCCGACG--- ((((((........(((....)))(((((((..(((....))(((((((((((......))))))))))))..)))))))))))))..--- ( -40.60, z-score = -2.98, R) >droSim1.chr3L 6143986 88 - 22553184 UCGGCGUGACCAGAAUGCUAGCAUACUUUUGAGCGCCGGAGCGGUGACCGCAGAAUGCGCUGCGGUCGCCGAGCAGAAGUUGCCGACG--- ((((((........(((....)))(((((((..(((....))(((((((((((......))))))))))))..)))))))))))))..--- ( -40.60, z-score = -2.98, R) >consensus UCGGCGUGACCAGAAUGCUAGCAUACUUUUCAGCGCCGGAGCGGUGACCGCAGAAUGCGCUGCGGUCGCCGAGCAGAAGUUGCCGACG___ (((((..(((.....((((.((..........)).......((((((((((((......))))))))))))))))...))))))))..... (-24.42 = -26.20 + 1.78)
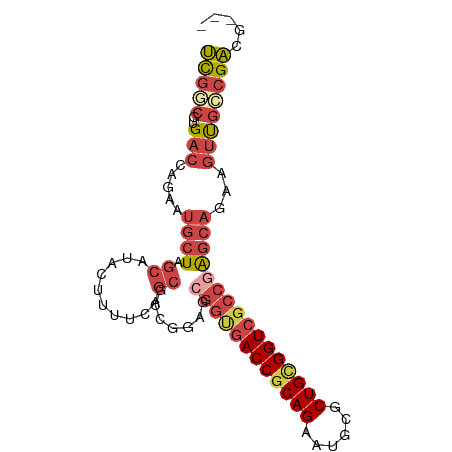
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:40 2011