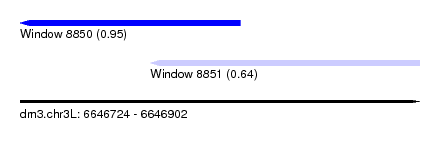
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,646,724 – 6,646,902 |
| Length | 178 |
| Max. P | 0.951754 |

| Location | 6,646,724 – 6,646,822 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 67.88 |
| Shannon entropy | 0.59783 |
| G+C content | 0.50675 |
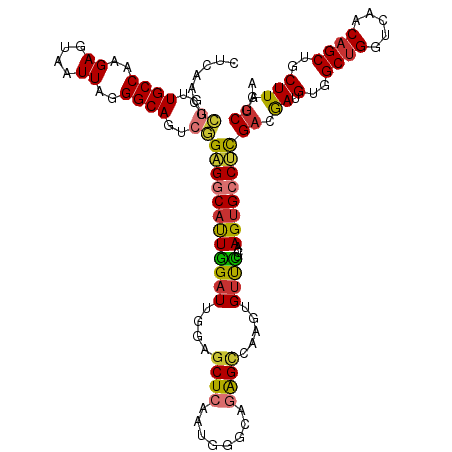
| Mean single sequence MFE | -31.41 |
| Consensus MFE | -16.56 |
| Energy contribution | -15.87 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
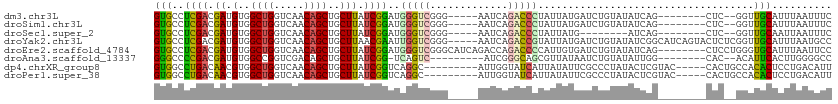
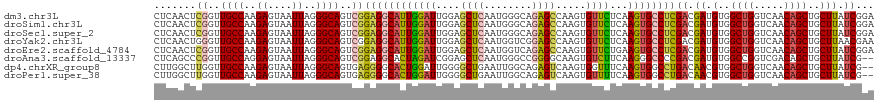
>dm3.chr3L 6646724 98 - 24543557 GUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAUCGGAUGGGUCGGG-----AAUCAGACCCUAUUAUGAUCUGUAUAUCAG--------CUC--GGUUGCAUUUAAUUUC ((((.((((..(.(..((((.....))))..))..))))..(((((...-----.....))))).....(((.(((.....)))--------.))--)...))))........ ( -28.80, z-score = -0.93, R) >droSim1.chr3L 6124704 98 - 22553184 GUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAUCGGAUGGGUCGGG-----AAUCAGACCCUAUUAUGAUCUGUAUAUCAG--------CUC--GGUUGCAUUUAAUUUC ((((.((((..(.(..((((.....))))..))..))))..(((((...-----.....))))).....(((.(((.....)))--------.))--)...))))........ ( -28.80, z-score = -0.93, R) >droSec1.super_2 6583554 90 - 7591821 GUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAUCGGAUGGGUCGGG-----AAUCAGACCCUAUUAUG--------AUCAG--------CUC--GGUUGCAAUUAAUUUC .(((.((((......(((((.....)))))(((.((((...(((((...-----.....))))).....))--------)).))--------)))--))..)))......... ( -27.20, z-score = -0.95, R) >droYak2.chr3L 7221285 108 - 24197627 GUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAACGAAUUGGUCGGG-----AAUCAGACCGUAUUAUGAUCUGUAUAUCGGCAUCAGUACUCUCGGUUGCAUUUAAUGCC ((((((((((.(.(..((((.....))))..).........).))))))-----.....(((((.....(((((((.....))).)))).......)))))))))........ ( -30.40, z-score = -0.55, R) >droEre2.scaffold_4784 21042757 105 + 25762168 GUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAUCGGAUGGGUCGGGCAUCAGACCAGACCCCAUUGUGAUCUGUAUAUCAG--------CUCCUGGGUGCAUUUAAUUCC (..(((.(.(((((..((((.....))))..)..))))((((((((((........)).)).))))))..((.(((.....)))--------.))).)))..).......... ( -33.90, z-score = -0.27, R) >droAna3.scaffold_13337 3006919 93 + 23293914 GGGCCCCGACGAUGUGGCCGGUCGACAGCUGCUUAUCGG-UCAGUC---------AUCGGGCAGCGUUAUAAUCUGUAUAUUGG--------CAC--ACAUUCACUUGGGGCC .((((((((.((((((((((((..(((((((((...(((-(.....---------))))))))))(.......))))..)))))--------).)--)))))...)))))))) ( -39.40, z-score = -3.05, R) >dp4.chrXR_group8 3919478 99 + 9212921 GUGGCCUGACAACGUGGCUGGUCAACAGCUGCUUAUCGGUCAGGC---------AUUGGUAUCAUUAUAUUCGCCCUAUACUCGUAC-----CACUGCCACACUCCUGACAUU ((((((((((...(..((((.....))))..)......)))))))---------..((((((.(.((((.......)))).).))))-----))....)))............ ( -31.40, z-score = -2.42, R) >droPer1.super_38 398628 99 - 801819 GUGGCCUGACAACGUGGCUGGUCAACAGCUGCUUAUCGGUCAGGC---------AUUGGUAUCAUUAUAUUCGCCCUAUACUCGUAC-----CACUGCCACACUCCUGACAUU ((((((((((...(..((((.....))))..)......)))))))---------..((((((.(.((((.......)))).).))))-----))....)))............ ( -31.40, z-score = -2.42, R) >consensus GUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAUCGGAUGGGUCGGG_____AAUCAGACCCUAUUAUGAUCUGUAUAUCAG________CUC__GGUUGCAUUUAAUUUC (((..((((.((.(..((((.....))))..))).))))..(((((.............)))))....................................))).......... (-16.56 = -15.87 + -0.69)
| Location | 6,646,782 – 6,646,902 |
|---|---|
| Length | 120 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.64 |
| Shannon entropy | 0.22422 |
| G+C content | 0.54100 |
| Mean single sequence MFE | -45.19 |
| Consensus MFE | -33.56 |
| Energy contribution | -33.24 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
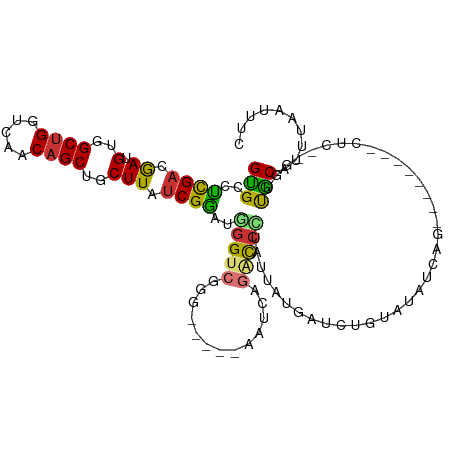
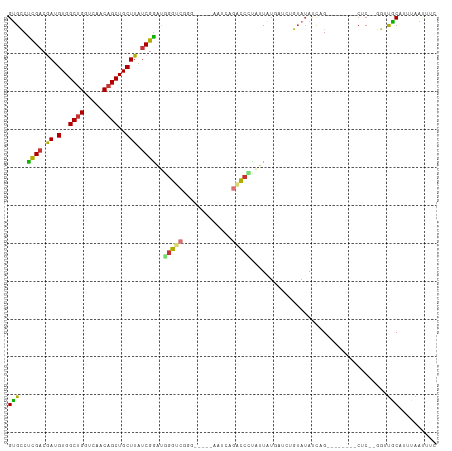
>dm3.chr3L 6646782 120 - 24543557 CUCAACUCGGUUGCCAAGAGUAAUUAGGGCAGUCGGAGGCAUUGGAUUGGAGCUCAAUGGGCAGAGCCAAGUGUUCUCAAGUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAUCGGA .......((..((((..((....))..))))..))((((((((......(((..((...(((...)))...))..))).))))))))..(((((..((((.....))))..)..)))).. ( -45.80, z-score = -2.03, R) >droSim1.chr3L 6124762 120 - 22553184 CUCAACUCGGUUGCCAAGAGUAAUUAGGGCAGUCGGAGGCAUUGGAUUGGAGCUCAAUGGGCAGAGCCAAGUGUUCUCAAGUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAUCGGA .......((..((((..((....))..))))..))((((((((......(((..((...(((...)))...))..))).))))))))..(((((..((((.....))))..)..)))).. ( -45.80, z-score = -2.03, R) >droSec1.super_2 6583604 120 - 7591821 CUCAACUCGGUUGCCAAGAGUAAUUAGGGCAGUCGGAGGCAUUGGAUUGGAGCUCAAUGGGCAGAGCCAAGUGUUCUCAAGUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAUCGGA .......((..((((..((....))..))))..))((((((((......(((..((...(((...)))...))..))).))))))))..(((((..((((.....))))..)..)))).. ( -45.80, z-score = -2.03, R) >droYak2.chr3L 7221353 120 - 24197627 CUCAACUGGGUUGCCAAGAGUAAUUAGGGCAGUCGGAGGCAUUGGAUUGGAGCUCAAUGGUCGGAGCCAAGUGUUCUCAAGUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAACGAA (((....))).((((..((....))..))))((((.(((((((.....(((((....((((....))))...)))))..)))))))))))...(..((((.....))))..)........ ( -40.80, z-score = -0.92, R) >droEre2.scaffold_4784 21042822 120 + 25762168 CUCAACUCGGUUGCCAAGAGUAAUUAGGGCAGUCGGAGGCAUUGGAUUGGAGCUCAAUGGUCAGAGCCAAGUGUUCUGAAGUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAUCGGA .......((..((((..((....))..))))..))((((((((...(..((((....((((....))))...))))..)))))))))..(((((..((((.....))))..)..)))).. ( -46.80, z-score = -2.47, R) >droAna3.scaffold_13337 3006974 118 + 23293914 CUCAGCCCGGUUGCCAGGAGUAAUUAGGGCAGUCGGAGGCACUAGAUCGGAGCUCAAUGGGCCGGGGCAAGUGUCUUCAAGGGCCCCGACGAUGUGGCCGGUCGACAGCUGCUUAUCG-- ..(((((((..((((..((....))..))))..))).(((.(((.((((..(((.....)))((((((...((....))...)))))).)))).)))...)))....)))).......-- ( -48.90, z-score = -1.27, R) >dp4.chrXR_group8 3919539 118 + 9212921 CUUGGCUUGGUUGCCAAGAGUAAUUAGGGCAGUGAGGGGCACUGGAUUGGGGCUGAAUUGGCAGAGUCAAGUGGUUUCAAGUGGCCUGACAACGUGGCUGGUCAACAGCUGCUUAUCG-- ((((((......)))))).((..(((((.((.(..(((.((((.((((...(((.....)))..)))).)))).)))..).)).)))))..))(..((((.....))))..)......-- ( -42.80, z-score = -1.62, R) >droPer1.super_38 398689 118 - 801819 CUUGGCUUGGUUGCCAAGAGUAAUUAGGGCAGUGAGGGGCACUGGAUUGGGGCUGAAUUGGCAGAGUCAAGUGUUUUCAAGUGGCCUGACAACGUGGCUGGUCAACAGCUGCUUAUCG-- ((((((......)))))).((..(((((.((.(..((((((((.((((...(((.....)))..)))).))))))))..).)).)))))..))(..((((.....))))..)......-- ( -44.80, z-score = -2.30, R) >consensus CUCAACUCGGUUGCCAAGAGUAAUUAGGGCAGUCGGAGGCAUUGGAUUGGAGCUCAAUGGGCAGAGCCAAGUGUUCUCAAGUGCCUCGACGAUGUGGCUGGUCAACAGCUGCUUAUCGGA .......((..((((..((....))..))))..))((((((((((((....((((........)))).....))))...))))))))((.((.(..((((.....))))..))).))... (-33.56 = -33.24 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:36 2011