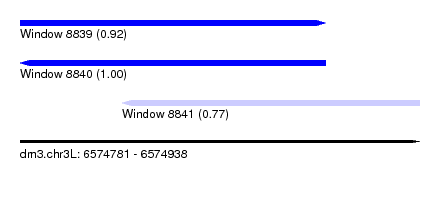
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,574,781 – 6,574,938 |
| Length | 157 |
| Max. P | 0.997315 |

| Location | 6,574,781 – 6,574,901 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 56.60 |
| Shannon entropy | 0.72388 |
| G+C content | 0.32911 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -6.34 |
| Energy contribution | -11.90 |
| Covariance contribution | 5.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6574781 120 + 24543557 UUAUUUCAAACUUACAAAGUGAGAGAUUUCAAUAAAAAAAGUUGAAACUAACAGUGAAAUCUCUGAGGUGUUAAGCAACUUUUACGAAAGGGCCUUAACACCCUUGGGAUUUCUUUGUUA ..........(((((...)))))((.(((((((.......)))))))))(((((.((((((((...(((((((((...((((....))))...)))))))))...)))))))).))))). ( -34.80, z-score = -3.86, R) >droEre2.scaffold_4784 9248758 93 + 25762168 -UAGUUUUAGUUCACAGAA--AAAGGUAAACAUAGGAAGA-UUCAGGAUAACACUGAAAACUGUAGGAUGUUGAGAAACUAAAACACCUCGUUGUUA----------------------- -..((((((((((((....--....)).(((((....((.-(((((.......)))))..)).....)))))..).)))))))))............----------------------- ( -15.30, z-score = 0.05, R) >droSec1.super_2 6511418 117 + 7591821 -UAGUUUCAAAUAACAGAAAGAG-AAUUUCAAUGAAAAUA-UUCAUAUGAACUCUAAAAUCUGUGGGGUGUUAAGAUACUUCCACCAAAGGGUCUUAACACCCUUGGGAUUUCUUUGUUA -..........(((((((.((((-...(((((((((....-))))).)))))))).((((((..((((((((((((..(((......)))..))))))))))))..)))))).))))))) ( -37.20, z-score = -4.57, R) >droSim1.chr3L 6046467 96 + 22553184 -----------------------UAGUUUCAAUAAAAAGA-UUCAGAUUAACACUAAAAUCUGGUGGGUGUUAAGAUACUCUUACCAAAGGGCCUUAACAACCUAGGGAUUUCUUUGUUA -----------------------.......((((((.(((-((((((((........)))))..(((((((((((...(((((....))))).)))))).))))).)))))).)))))). ( -22.30, z-score = -1.41, R) >consensus _UAGUUU_AA_U_ACAGAA__AGAAAUUUCAAUAAAAAGA_UUCAGAUUAACACUAAAAUCUGUGGGGUGUUAAGAAACUUUUACCAAAGGGCCUUAACACCCUUGGGAUUUCUUUGUUA ........................................................((((((..(((((((((((((.(((......))).)))))))))))))..))))))........ ( -6.34 = -11.90 + 5.56)

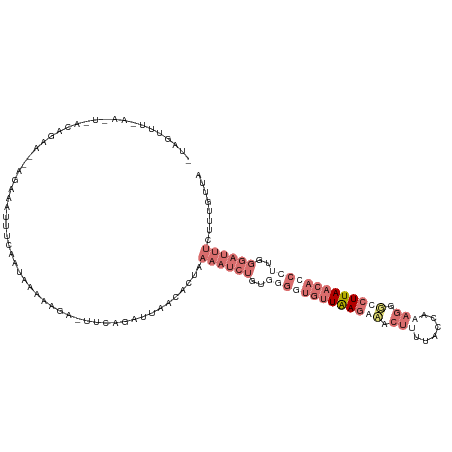
| Location | 6,574,781 – 6,574,901 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 56.60 |
| Shannon entropy | 0.72388 |
| G+C content | 0.32911 |
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -10.00 |
| Energy contribution | -12.12 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.997315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6574781 120 - 24543557 UAACAAAGAAAUCCCAAGGGUGUUAAGGCCCUUUCGUAAAAGUUGCUUAACACCUCAGAGAUUUCACUGUUAGUUUCAACUUUUUUUAUUGAAAUCUCUCACUUUGUAAGUUUGAAAUAA (((((..((((((.(..((((((((((...((((....))))...))))))))))..).))))))..)))))((((((((((.......(((......)))......))).))))))).. ( -30.42, z-score = -3.26, R) >droEre2.scaffold_4784 9248758 93 - 25762168 -----------------------UAACAACGAGGUGUUUUAGUUUCUCAACAUCCUACAGUUUUCAGUGUUAUCCUGAA-UCUUCCUAUGUUUACCUUU--UUCUGUGAACUAAAACUA- -----------------------............((((((((((..((((((.....((..(((((.(....))))))-.))....))))........--...)).))))))))))..- ( -12.00, z-score = 0.15, R) >droSec1.super_2 6511418 117 - 7591821 UAACAAAGAAAUCCCAAGGGUGUUAAGACCCUUUGGUGGAAGUAUCUUAACACCCCACAGAUUUUAGAGUUCAUAUGAA-UAUUUUCAUUGAAAUU-CUCUUUCUGUUAUUUGAAACUA- ...((((..........(((((((((((..((((....))))..))))))))))).(((((....((((((((.(((((-....)))))))))...-)))).)))))..))))......- ( -34.40, z-score = -4.24, R) >droSim1.chr3L 6046467 96 - 22553184 UAACAAAGAAAUCCCUAGGUUGUUAAGGCCCUUUGGUAAGAGUAUCUUAACACCCACCAGAUUUUAGUGUUAAUCUGAA-UCUUUUUAUUGAAACUA----------------------- (((((..((((((....((.((((((((..((((....))))..)))))))).))....))))))..))))).......-.................----------------------- ( -22.00, z-score = -2.09, R) >consensus UAACAAAGAAAUCCCAAGGGUGUUAAGACCCUUUGGUAAAAGUAUCUUAACACCCCACAGAUUUCAGUGUUAAUCUGAA_UCUUUUUAUUGAAACCUCU__UUCUGU_A_UU_AAACUA_ .................(((((((((((..((((....))))..)))))))))))..((((((........))))))........................................... (-10.00 = -12.12 + 2.12)

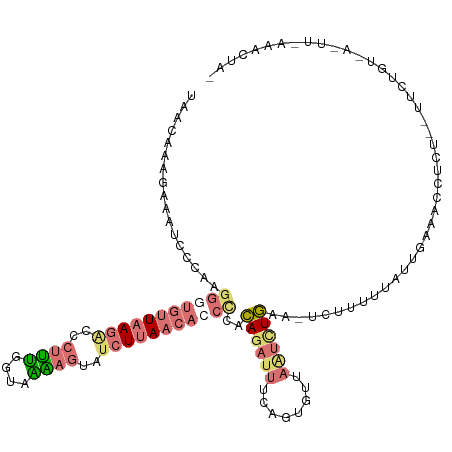
| Location | 6,574,821 – 6,574,938 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 73.53 |
| Shannon entropy | 0.43757 |
| G+C content | 0.35033 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -16.31 |
| Energy contribution | -19.38 |
| Covariance contribution | 3.06 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.767484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6574821 117 - 24543557 AUCUUGUAAACAACUACUAAAUAGAACUUGAAAGAGGUAACAAAGAAAUCCCAAGGGUGUUAAGGCCCUUUCGUAAAAGUUGCUUAACACCUCAGAGAUUUCACUGUUAGUUUCAAC ...(((.((((...............(((....))).(((((..((((((.(..((((((((((...((((....))))...))))))))))..).))))))..)))))))))))). ( -29.90, z-score = -2.60, R) >droEre2.scaffold_4784 9248795 93 - 25762168 AUCUUUUAAACAACUUCAGAAUGGAUCAUGAAAAGGGUAACAACGA---------GGUGU--------UUU------AGUUUCUCAACAUCCUACAGUUUUCAGUGUUAUCCUGAA- ..............((((((.....)).)))).(((((((((((.(---------(((((--------(..------........)))).)))...))......)))))))))...- ( -14.70, z-score = 0.70, R) >droSec1.super_2 6511456 116 - 7591821 AUCUUGUAAACAACUUCUAAAUAGAACUUGAAAAGGGUAACAAAGAAAUCCCAAGGGUGUUAAGACCCUUUGGUGGAAGUAUCUUAACACCCCACAGAUUUUAGAGUUCAUAUGAA- .......................((((((.(((((((............)))..(((((((((((..((((....))))..)))))))))))......)))).)))))).......- ( -31.20, z-score = -2.41, R) >droSim1.chr3L 6046484 116 - 22553184 AUAUUGUAAACCACUUCUAAAAAGAACUUGAAAAGGGUAACAAAGAAAUCCCUAGGUUGUUAAGGCCCUUUGGUAAGAGUAUCUUAACACCCACCAGAUUUUAGUGUUAAUCUGAA- ......................(((.(((....))).(((((..((((((....((.((((((((..((((....))))..)))))))).))....))))))..))))).)))...- ( -25.00, z-score = -0.96, R) >consensus AUCUUGUAAACAACUUCUAAAUAGAACUUGAAAAGGGUAACAAAGAAAUCCCAAGGGUGUUAAGGCCCUUUGGUAAAAGUAUCUUAACACCCCACAGAUUUCAGUGUUAAUCUGAA_ .................................(((.(((((..((((((....(((((((((((..((((....))))..)))))))))))....))))))..))))).))).... (-16.31 = -19.38 + 3.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:28 2011