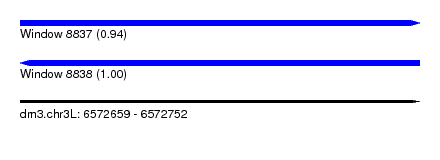
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,572,659 – 6,572,752 |
| Length | 93 |
| Max. P | 0.998608 |

| Location | 6,572,659 – 6,572,752 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 74.52 |
| Shannon entropy | 0.49938 |
| G+C content | 0.38389 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -9.45 |
| Energy contribution | -10.07 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
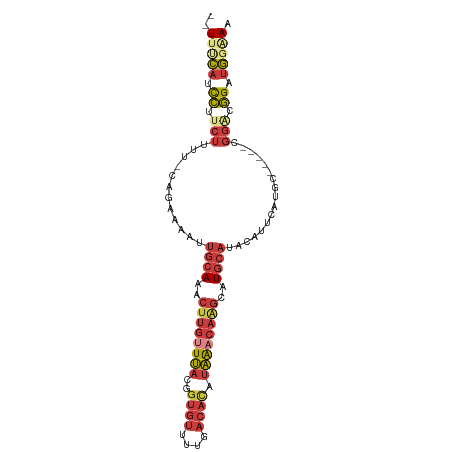
>dm3.chr3L 6572659 93 + 24543557 UUUUUCAUCCUUCUUUU-CAGAAAAUUGCAAACUUGUUUACGGUGUUUUGACACAUAAACAAGCAUGCAUACAUUCAUGC-----CGGACGGGUGGGAA .((..(((((.(((...-(((((...((((..((((((((..((((....)))).))))))))..))))....))).)).-----.))).)))))..)) ( -28.20, z-score = -3.77, R) >droAna3.scaffold_13337 2929261 88 - 23293914 --UUUUUGCCGUCUUUU-GAGAGAAUUGCAAACUUGUUUACGGUGUUUUGACACAUAA-CAUGCAUGCAAACAUAUAGG-------GGAAGGAUAUAAA --(((((.((.((((..-..)))).(((((..(.((((....((((....))))..))-)).)..))))).......))-------.)))))....... ( -17.30, z-score = -0.80, R) >droEre2.scaffold_4784 9246708 95 + 25762168 --UUUCAUCCUUCUUUU-CAGAAAAUUGCAAACUUGUUUACGGUGUUUUGACACAUAAACAAGCAUGCAUACAUUCAUGCGAUUCAGGGCGGCUGGAA- --.((((.((.......-..(((...((((..((((((((..((((....)))).))))))))..))))....)))..((........)))).)))).- ( -22.50, z-score = -1.65, R) >droYak2.chr3L 7144338 91 + 24197627 --UUUCAUCCUGCUUUU-CAGAAAAUUGCAAACUUGUUUACGCUGUUUUGACACAUAAACAAGCAUGCAUACAUUCAUGC-----AGGGCGGAUGGUAA --..((((((.(((((.-(((((...((((..((((((((.(.(((....)))).))))))))..))))....))).)).-----)))))))))))... ( -27.70, z-score = -3.51, R) >droSec1.super_2 6509320 91 + 7591821 --UUUCAUCCUUCUUUU-CAGAAAAUUGCAAACUUGUUUACGGUGUUUUGACACAUAAACAAGCAUGCAUACAUUCAUGU-----CGGACGGGUGGAAA --((((((((.(((...-(((((...((((..((((((((..((((....)))).))))))))..))))....))).)).-----.))).)))))))). ( -28.80, z-score = -4.38, R) >droSim1.chr3L 6044355 93 + 22553184 UUUUCCAUCCUUCUUUU-CAGAAAAUUGCAAACUUGUUUACGGUGUUUUGACACAUAAACAAGCAUGCAUACAUUCAUGC-----CGGACGGGUGGGAA .(((((((((.(((...-(((((...((((..((((((((..((((....)))).))))))))..))))....))).)).-----.))).))))))))) ( -30.80, z-score = -4.32, R) >droGri2.scaffold_15110 16180349 87 - 24565398 -UUUGCACUGUCCAUAUGCAUGUGAAGGGAGACUUUUUGCUGGUCUUUGUUUUUCUUGUUUGCCAUGGAGUGAUUCA----------AAAUGAUGGCA- -...(((..(.(((...(((...(((((....))))))))))).)..)))..........(((((((((....))))----------......)))))- ( -19.00, z-score = 0.41, R) >consensus __UUUCAUCCUUCUUUU_CAGAAAAUUGCAAACUUGUUUACGGUGUUUUGACACAUAAACAAGCAUGCAUACAUUCAUGC_____CGGACGGAUGGAAA ..(((((.((...(((....)))...((((..((((((((..((((....)))).))))))))..)))).....................)).))))). ( -9.45 = -10.07 + 0.62)

| Location | 6,572,659 – 6,572,752 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 74.52 |
| Shannon entropy | 0.49938 |
| G+C content | 0.38389 |
| Mean single sequence MFE | -24.71 |
| Consensus MFE | -12.15 |
| Energy contribution | -15.13 |
| Covariance contribution | 2.98 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.998608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
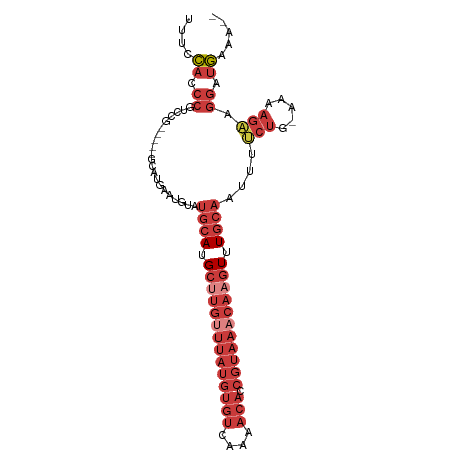
>dm3.chr3L 6572659 93 - 24543557 UUCCCACCCGUCCG-----GCAUGAAUGUAUGCAUGCUUGUUUAUGUGUCAAAACACCGUAAACAAGUUUGCAAUUUUCUG-AAAAGAAGGAUGAAAAA ........(((((.-----...........((((.((((((((((((((....))).))))))))))).))))...((((.-...)))))))))..... ( -29.90, z-score = -4.78, R) >droAna3.scaffold_13337 2929261 88 + 23293914 UUUAUAUCCUUCC-------CCUAUAUGUUUGCAUGCAUG-UUAUGUGUCAAAACACCGUAAACAAGUUUGCAAUUCUCUC-AAAAGACGGCAAAAA-- .......((.((.-------.........(((((.((.((-((((((((....))).))).)))).)).))))).......-....)).))......-- ( -12.30, z-score = -0.64, R) >droEre2.scaffold_4784 9246708 95 - 25762168 -UUCCAGCCGCCCUGAAUCGCAUGAAUGUAUGCAUGCUUGUUUAUGUGUCAAAACACCGUAAACAAGUUUGCAAUUUUCUG-AAAAGAAGGAUGAAA-- -.(((....((........)).........((((.((((((((((((((....))).))))))))))).))))...((((.-...))))))).....-- ( -26.30, z-score = -3.19, R) >droYak2.chr3L 7144338 91 - 24197627 UUACCAUCCGCCCU-----GCAUGAAUGUAUGCAUGCUUGUUUAUGUGUCAAAACAGCGUAAACAAGUUUGCAAUUUUCUG-AAAAGCAGGAUGAAA-- ....(((((((...-----.((.(((.((.((((.((((((((((((((....))).))))))))))).)))))).)))))-....)).)))))...-- ( -30.50, z-score = -4.39, R) >droSec1.super_2 6509320 91 - 7591821 UUUCCACCCGUCCG-----ACAUGAAUGUAUGCAUGCUUGUUUAUGUGUCAAAACACCGUAAACAAGUUUGCAAUUUUCUG-AAAAGAAGGAUGAAA-- ........(((((.-----...........((((.((((((((((((((....))).))))))))))).))))...((((.-...)))))))))...-- ( -29.90, z-score = -5.26, R) >droSim1.chr3L 6044355 93 - 22553184 UUCCCACCCGUCCG-----GCAUGAAUGUAUGCAUGCUUGUUUAUGUGUCAAAACACCGUAAACAAGUUUGCAAUUUUCUG-AAAAGAAGGAUGGAAAA .......((((((.-----...........((((.((((((((((((((....))).))))))))))).))))...((((.-...)))))))))).... ( -33.20, z-score = -5.20, R) >droGri2.scaffold_15110 16180349 87 + 24565398 -UGCCAUCAUUU----------UGAAUCACUCCAUGGCAAACAAGAAAAACAAAGACCAGCAAAAAGUCUCCCUUCACAUGCAUAUGGACAGUGCAAA- -((((.((....----------.)).....(((((((((.....(((......((((.........))))...)))...))).))))))..).)))..- ( -10.90, z-score = 0.83, R) >consensus UUUCCACCCGUCCG_____GCAUGAAUGUAUGCAUGCUUGUUUAUGUGUCAAAACACCGUAAACAAGUUUGCAAUUUUCUG_AAAAGAAGGAUGAAA__ ....((.((.....................((((.((((((((((((((....))).))))))))))).))))....(((.....))).)).))..... (-12.15 = -15.13 + 2.98)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:26 2011