
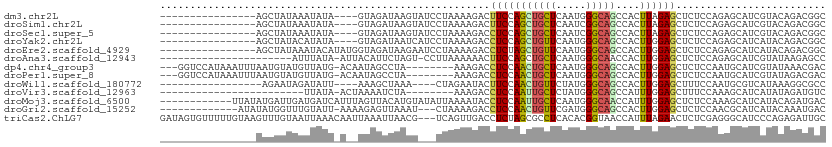
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,453,264 – 4,453,355 |
| Length | 91 |
| Max. P | 0.638268 |

| Location | 4,453,264 – 4,453,355 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 64.04 |
| Shannon entropy | 0.74456 |
| G+C content | 0.42033 |
| Mean single sequence MFE | -19.69 |
| Consensus MFE | -8.66 |
| Energy contribution | -8.32 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.64 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
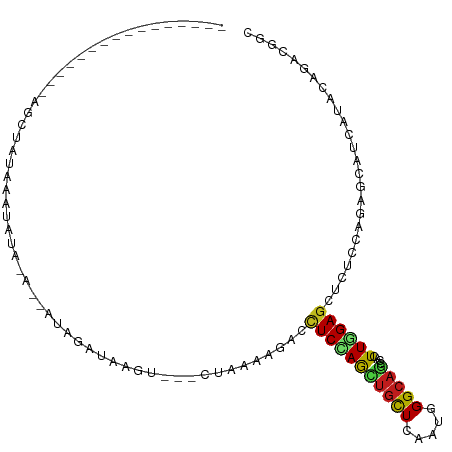
>dm3.chr2L 4453264 91 + 23011544 ----------------AGCUAUAAAUAUA----GUAGAUAAGUAUCCUAAAAGACUUCCAGCUGCUCAAUGGGCAGCCACUUAGAGCUCUCCAGAGCAUCGUACAGACGGC ----------------.(((((....)))----))......((((...............((((((.....))))))......((((((....)))).))))))....... ( -21.30, z-score = -0.46, R) >droSim1.chr2L 4379630 91 + 22036055 ----------------AGCUAUAAAUAUA----GUAGAUAAGUAUCCUAAAAGACUUCCAGCUGCUCAAUCGGCAGCCACUUAGAGCUCUCCAGAGCAUCGUACAGACGGC ----------------.(((((....)))----))......((((...............((((((.....))))))......((((((....)))).))))))....... ( -19.70, z-score = -0.41, R) >droSec1.super_5 2551128 91 + 5866729 ----------------AGCUAUAAAUAUA----GUAGAUAAGUAUCCUAAAAGACCUCCAGCUGCUCAAUCGGCAGCCACUUAGAGCUCUCCAGAGCAUCGUACAGACGGC ----------------.(((((....)))----))......((((...............((((((.....))))))......((((((....)))).))))))....... ( -19.70, z-score = -0.43, R) >droYak2.chr2L 4488304 91 + 22324452 ----------------AGCUAUACAUAUA----GUAGAUAAUCAUCCUAAAAGACCUCCAGCUGUUCAAUGGGCAGCCACUUGGAGCUCUCCAGAGCAUCAUACAGACGGC ----------------.(((((....)))----)).(((.................((((((((((.....))))))....))))((((....)))))))........... ( -20.40, z-score = -0.42, R) >droEre2.scaffold_4929 4540481 95 + 26641161 ----------------AGCUAUAAAUACAUAUGGUAGAUAAGAAUCCUAAAAGACCUCUAGCUGUUCAAUGGGCAGCCACUUGGAGCUCUCCAGAGCAUCAUACAGACGGC ----------------.((((((......))))))...................(((((.((((((.....))))))....((..((((....))))..))...))).)). ( -20.90, z-score = -0.04, R) >droAna3.scaffold_12943 1647016 87 - 5039921 ----------------------AUUUAUA-AUUACAUUCUAGU-CCUUAAAAAACUUCCAGCUGCUCAAUGGGCAACCACUUGGAGCUCUCCAGAGCAUCGUAUAAGAGCC ----------------------.......-.............-..........(((...(.(((((..(((....)))..((((....))))))))).)....))).... ( -17.40, z-score = -0.95, R) >dp4.chr4_group3 2403177 99 - 11692001 ---GGUCCAUAAAUUUAAUGUAUGUUAUG-ACAAUAGCCUA--------AAAGACCUCCAACUGCUCAAUGGGCAGCCACUUGGAGCUCUCCAAUGCAUCGUAUAAACGAC ---...............((((((((((.-...)))))...--------..(((.(((((((((((.....)))))....)))))).)))...)))))((((....)))). ( -23.10, z-score = -1.68, R) >droPer1.super_8 3486868 99 - 3966273 ---GGUCCAUAAAUUUAAUGUAUGUUAUG-ACAAUAGCCUA--------AAAGACCUCCAACUGCUCAAUGGGCAGCCACUUGGAGCUCUCCAAUGCAUCGUAUAGACGAC ---.(((.(((......(((((((((((.-...)))))...--------..(((.(((((((((((.....)))))....)))))).)))...))))))..))).)))... ( -24.60, z-score = -1.76, R) >droWil1.scaffold_180772 4881442 86 - 8906247 -----------------AGAAUAGAUAUU----AAAGCUAAA----CUAGAAUACUUCCAACUGUUCUAUGGGCAGCCACUUGGAGCUUUCCAAUGCGUCAUAAAGGCGCC -----------------............----...(((...----.(((((((........)))))))..)))......((((......)))).(((((.....))))). ( -17.50, z-score = -0.12, R) >droVir3.scaffold_12963 2491627 78 - 20206255 ------------------------UUAUA-ACUAAAAUCUA--------AAAGACCUCCAAUUGCUCUAUGGGCAGCCAUUUGGAGCUUUCCAAAGCAUCAUAUAGAUGUC ------------------------.....-...........--------((((..(((((((((((.....)))))....)))))))))).....(((((.....))))). ( -15.40, z-score = -1.13, R) >droMoj3.scaffold_6500 9598306 99 - 32352404 ------------UUAUAUGAUUGAUGAUCAUUUAGUUACAUGUAUAUUAAAAUACCUCCAAUUGCUCAAUGGGCAACCAUUUGGAGCUCUCCAAAGCAUCAUACAGAUGAC ------------.((.((((((...)))))).))((((..(((((...........(((((.......((((....)))))))))(((......)))...)))))..)))) ( -19.61, z-score = -0.92, R) >droGri2.scaffold_15252 2040415 94 - 17193109 -------------AUAUAUGGUUUGUAUU-AAAAGAGUUAAAU---CUAAAAGACCUCCAACUGUUCGAUGGGCAGCCACUUGGAGCUCUCCAACGCAUCAUACAAAUGAC -------------.....(.((((((((.-...(((......)---))...(((.(((((((((((.....)))))....)))))).)))..........)))))))).). ( -19.90, z-score = -1.17, R) >triCas2.ChLG7 12399695 108 - 17478683 GAUAGUGUUUUUGUAAGUUUGUAAUUAAACAAUUAAAUUAACG---UCAGUUGACCUCUAGCGCCUCACACGGUAACCAUUUAGAACUCUCGAGGGCAUCCCAGAGAUUGC ....(((((((((..(((((.................(((((.---...)))))........(((......))).........)))))..)))))))))............ ( -16.50, z-score = 1.17, R) >consensus ________________AGCUAUAAAUAUA_A__AUAGAUAAGU___CUAAAAGACCUCCAGCUGCUCAAUGGGCAGCCACUUGGAGCUCUCCAGAGCAUCAUACAGACGGC .......................................................(((((((((((.....)))))....))))))......................... ( -8.66 = -8.32 + -0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:30 2011