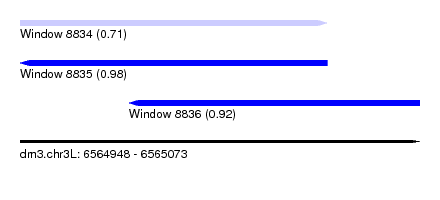
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,564,948 – 6,565,073 |
| Length | 125 |
| Max. P | 0.982814 |

| Location | 6,564,948 – 6,565,044 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 66.28 |
| Shannon entropy | 0.70498 |
| G+C content | 0.63490 |
| Mean single sequence MFE | -39.96 |
| Consensus MFE | -17.45 |
| Energy contribution | -17.38 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
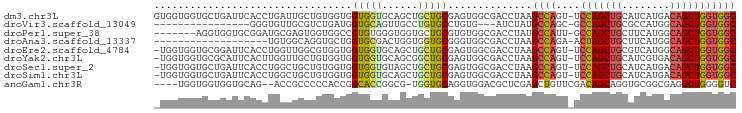
>dm3.chr3L 6564948 96 + 24543557 GUGGUGGUGCUGAUUCACCUGAUUGCUGUGGUGGUGGUGCAGCUGCUGCGAGUGGCGACCUAAGCCAGU-UCCAGCUGCAUCAUGACAGCUGGUGGC ..(((((.......)))))...(..((((.((.((((((((((((..((...((((.......))))))-..)))))))))))).)).)).))..). ( -40.00, z-score = -0.86, R) >droVir3.scaffold_13049 10922440 77 + 25233164 ----------------GGGUGUUGCGUCUGAUGGUGCAGUUGCCUGUGCCUGUG---AUCUAUGCCAGC-GCCAGCUGCGCCAUGGCAGCUGGUGGC ----------------.(((((...(((....((..(((....)))..))...)---))..)))))..(-(((((((((......)))))))))).. ( -36.00, z-score = -1.19, R) >droPer1.super_38 312347 89 + 801819 -------AGGUGGUGCGGAUGCGAGUGGUGGCCGUGUGGGUGGUGCUGCGUGUGGCGACCUAUGCCAUU-GCCAGCUGCUUCAUGGCAGCUGGUGGC -------.((((((((....))....(((.((((..((((.....)).))..)))).)))...))))))-((((((((((....))))))))))... ( -41.10, z-score = -1.26, R) >droAna3.scaffold_13337 2921279 77 - 23293914 -------------------UGUGGCAGGUGCUGGUGCGACUGGUGGUGCGGGUGGCGACCUAAGCCAGA-ACUAGCUGCUUCAUGGCAGCUGGUGGC -------------------....((.(((...(((((.(((.(.....).))).)).)))...)))...-((((((((((....)))))))))).)) ( -33.60, z-score = -1.21, R) >droEre2.scaffold_4784 9238601 95 + 25762168 -UGGUGGUGCGGAUUCACCUGGUUGGCGUGGUGGUGGUGCAGCUGCUGCGAGUGGCGACCUAAGCCAGU-UCCAGCUGCGUCAUGGCAGCUGGUGGC -.(((((.......))))).(((((.(((.(..(..(.....)..)..).).)).)))))...((((..-.((((((((......)))))))))))) ( -42.40, z-score = -0.73, R) >droYak2.chr3L 7135515 95 + 24197627 -UGGUGGUGCGCAUUCACUUGGUUGCUGUGGUGGUGGUGCAGCGGCUGCGAGUGGCGACCUAAGCCAGU-UCCAGCUGCAUCGUGACAGCUGGUGGC -....(((.(((...((((((((((((((.........))))))))..))))))))))))...((((..-.((((((((.....).))))))))))) ( -39.20, z-score = 0.00, R) >droSec1.super_2 6501418 95 + 7591821 -UGGUGGUGCUGAUUCACCUGGCUGCUGUGGUGGUGGUGUAGCUGCUGCGAGUGGCGACCUAAGCCAGU-UCCAGCUGCAUCAUGACAGCUGGUGGC -.(((((.......)))))..((..((((.((.((((((((((((..((...((((.......))))))-..)))))))))))).)).)).))..)) ( -41.60, z-score = -1.21, R) >droSim1.chr3L 6036468 95 + 22553184 -UGGUGGUGCUGAUUCACCUGGCUGCUGUGGUGGUGGUGCAGCUGCUGCGAGUGGCGACCUAAGCCAGU-UCCAGCUGCAUCAUGACAGCUGGUGGC -.(((((.......)))))..((..((((.((.((((((((((((..((...((((.......))))))-..)))))))))))).)).)).))..)) ( -43.60, z-score = -1.33, R) >anoGam1.chr3R 35999276 90 + 53272125 ----UGGUGGUGGUGCAG--ACCGCCCCCACCGGCACCGGCG-UGGUGCAGGUGGACGCUCGAGCUGUUCGACAGCAGGUGCGGCGAGGCUGGGGUC ----....((((((....--)))))).(((((.(((((....-.))))).)))))..((((.((((..(((...((....))..))))))).)))). ( -42.10, z-score = 0.10, R) >consensus _UGGUGGUGCUGAUUCACCUGGUUGCUGUGGUGGUGGUGCAGCUGCUGCGAGUGGCGACCUAAGCCAGU_UCCAGCUGCAUCAUGGCAGCUGGUGGC .................................(((((......)))))...((((.......))))....(((((((........))))))).... (-17.45 = -17.38 + -0.07)
| Location | 6,564,948 – 6,565,044 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 66.28 |
| Shannon entropy | 0.70498 |
| G+C content | 0.63490 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -15.60 |
| Energy contribution | -16.29 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.982814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
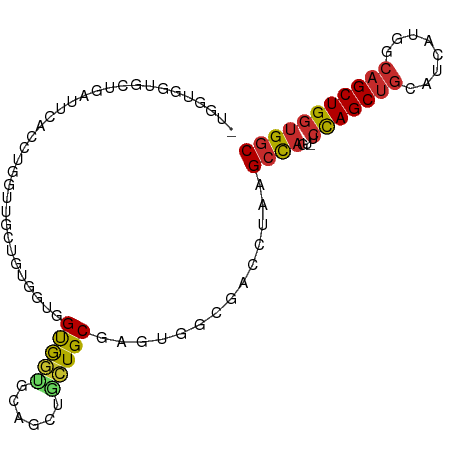
>dm3.chr3L 6564948 96 - 24543557 GCCACCAGCUGUCAUGAUGCAGCUGGA-ACUGGCUUAGGUCGCCACUCGCAGCAGCUGCACCACCACCACAGCAAUCAGGUGAAUCAGCACCACCAC ((((((((((((......)))))))).-..))))...(((.((.....((((...)))).....((((..........)))).....)))))..... ( -31.90, z-score = -1.22, R) >droVir3.scaffold_13049 10922440 77 - 25233164 GCCACCAGCUGCCAUGGCGCAGCUGGC-GCUGGCAUAGAU---CACAGGCACAGGCAACUGCACCAUCAGACGCAACACCC---------------- ((((((((((((......)))))))).-..))))......---....((..(((....)))..))................---------------- ( -28.50, z-score = -1.50, R) >droPer1.super_38 312347 89 - 801819 GCCACCAGCUGCCAUGAAGCAGCUGGC-AAUGGCAUAGGUCGCCACACGCAGCACCACCCACACGGCCACCACUCGCAUCCGCACCACCU------- ((((((((((((......)))))))).-..))))...(((.(((....................))).)))....((....)).......------- ( -29.45, z-score = -2.27, R) >droAna3.scaffold_13337 2921279 77 + 23293914 GCCACCAGCUGCCAUGAAGCAGCUAGU-UCUGGCUUAGGUCGCCACCCGCACCACCAGUCGCACCAGCACCUGCCACA------------------- (((((.((((((......)))))).))-...)))...(((.((.....)))))..(((..((....))..))).....------------------- ( -22.10, z-score = -0.82, R) >droEre2.scaffold_4784 9238601 95 - 25762168 GCCACCAGCUGCCAUGACGCAGCUGGA-ACUGGCUUAGGUCGCCACUCGCAGCAGCUGCACCACCACCACGCCAACCAGGUGAAUCCGCACCACCA- ((((((((((((......)))))))).-..))))...(((.((.....((((...))))..........((((.....)))).....)))))....- ( -34.50, z-score = -2.27, R) >droYak2.chr3L 7135515 95 - 24197627 GCCACCAGCUGUCACGAUGCAGCUGGA-ACUGGCUUAGGUCGCCACUCGCAGCCGCUGCACCACCACCACAGCAACCAAGUGAAUGCGCACCACCA- ((((((((((((......)))))))).-..))))...(((.((((.((((....((((...........))))......)))).)).)))))....- ( -32.40, z-score = -1.47, R) >droSec1.super_2 6501418 95 - 7591821 GCCACCAGCUGUCAUGAUGCAGCUGGA-ACUGGCUUAGGUCGCCACUCGCAGCAGCUACACCACCACCACAGCAGCCAGGUGAAUCAGCACCACCA- ((((((((((((......)))))))).-..))))...(((.((...((((.((.(((.............))).))...))))....)))))....- ( -30.22, z-score = -0.70, R) >droSim1.chr3L 6036468 95 - 22553184 GCCACCAGCUGUCAUGAUGCAGCUGGA-ACUGGCUUAGGUCGCCACUCGCAGCAGCUGCACCACCACCACAGCAGCCAGGUGAAUCAGCACCACCA- ((((((((((((......)))))))).-..))))...(((.((...((((....(((((............)))))...))))....)))))....- ( -34.70, z-score = -1.34, R) >anoGam1.chr3R 35999276 90 - 53272125 GACCCCAGCCUCGCCGCACCUGCUGUCGAACAGCUCGAGCGUCCACCUGCACCA-CGCCGGUGCCGGUGGGGGCGGU--CUGCACCACCACCA---- ((((...(((((..(((.(..((((.....))))..).)))..((((.(((((.-....))))).))))))))))))--).............---- ( -38.60, z-score = -0.91, R) >consensus GCCACCAGCUGCCAUGAUGCAGCUGGA_ACUGGCUUAGGUCGCCACUCGCAGCAGCUGCACCACCACCACAGCAACCAGGUGAAUCAGCACCACCA_ ((((((((((((......))))))))....))))............................................................... (-15.60 = -16.29 + 0.69)
| Location | 6,564,982 – 6,565,073 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 73.67 |
| Shannon entropy | 0.50784 |
| G+C content | 0.59055 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.17 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6564982 91 - 24543557 CGCACAAAUCCGUUUCCCAUCUCGACUUGGCCACCAGCU--GUCAUGAUGCAGCUGGAACUGGCUUAGGUCGCCACUCGCA-GCAGCUGCACCA------------------ ......................(((((((((((((((((--((......))))))))...)))).)))))))......(((-(...))))....------------------ ( -30.70, z-score = -1.49, R) >droGri2.scaffold_15110 16171734 109 + 24565398 CGCACAAAUCCGUAUCGCAUCUCGAUUUGGCCACCAGCU--GCCAUGGUGCAGCUGGCGCUGGCAUAGAUCGGCAACGCAG-GCAACUGCAAGUGCAACUGCAUCAUCAUCA .((((...........((....(((((((((((((((((--((......))))))))...)))).)))))))))...((((-....))))..))))................ ( -43.80, z-score = -2.58, R) >droVir3.scaffold_13049 10922458 88 - 25233164 CGCACAAAUCCGUUUCGCAUCUCGAUUUGGCCACCAGCU--GCCAUGGCGCAGCUGGCGCUGGCAUAGAUCACAGGCACAG-GCAACUGCA--------------------- ................((.....((((((((((((((((--((......))))))))...)))).))))))....)).(((-....)))..--------------------- ( -31.80, z-score = -0.98, R) >droMoj3.scaffold_6680 14861785 91 + 24764193 CGCACAAAUCCGUCUCGCAUCUCGAUUUGGCCACCAGCU--GCCAUGGCGCAGCUGGCGCUGGCAUAGAUCACAGGCAGAG-GCAACUGCUGCA------------------ .(((((.....(((((((.....((((((((((((((((--((......))))))))...)))).))))))....)).)))-))...)).))).------------------ ( -36.50, z-score = -1.26, R) >droPer1.super_38 312380 85 - 801819 CGCACAAAUCCGUGUCGCAUCUCGAUUUGGCCACCAGCU--GCCAUGAAGCAGCUGGCAAUGGCAUAGGUCGCCACACGCA-GCACCA------------------------ ...........((((.((....(((((((((((((((((--((......))))))))...)))).)))))))......)).-))))..------------------------ ( -32.50, z-score = -2.17, R) >dp4.chrXR_group8 3832107 85 + 9212921 CGCACAAAUCGGUGUCGCAUCUCGAUUUGGCCACCAGCU--GCCAUGAAGCAGCUGGCAAUGGCAUAGGUCGCCACACGCA-GCACCA------------------------ ..........(((((.((....(((((((((((((((((--((......))))))))...)))).)))))))......)).-))))).------------------------ ( -36.70, z-score = -3.23, R) >droAna3.scaffold_13337 2921294 91 + 23293914 CCCACAAAUCCGUCUCGCAUCUCGACUUGGCCACCAGCU--GCCAUGAAGCAGCUAGUUCUGGCUUAGGUCGCCACCCGCA-CCACCAGUCGCA------------------ ................((..((.(((((((((((.((((--((......)))))).))...))).))))))((.....)).-.....))..)).------------------ ( -24.90, z-score = -1.20, R) >droEre2.scaffold_4784 9238634 91 - 25762168 CACACAAAUCCGUUUCCCAUCUCGACUUGGCCACCAGCU--GCCAUGACGCAGCUGGAACUGGCUUAGGUCGCCACUCGCA-GCAGCUGCACCA------------------ ......................(((((((((((((((((--((......))))))))...)))).)))))))......(((-(...))))....------------------ ( -32.50, z-score = -2.64, R) >droYak2.chr3L 7135548 91 - 24197627 CACACAAAUCCGUUUCCCAUCUCGACUUGGCCACCAGCU--GUCACGAUGCAGCUGGAACUGGCUUAGGUCGCCACUCGCA-GCCGCUGCACCA------------------ ......................(((((((((((((((((--((......))))))))...)))).)))))))......(((-(...))))....------------------ ( -30.70, z-score = -2.22, R) >droSec1.super_2 6501451 91 - 7591821 CGCACAAAUCCGUUUCCCAUCUCGACUUGGCCACCAGCU--GUCAUGAUGCAGCUGGAACUGGCUUAGGUCGCCACUCGCA-GCAGCUACACCA------------------ .((....................((((((((((((((((--((......))))))))...)))).))))))((.....)).-))..........------------------ ( -29.30, z-score = -1.75, R) >droSim1.chr3L 6036501 91 - 22553184 CGCACAAAUCCGUUUCCCAUCUCGACUUGGCCACCAGCU--GUCAUGAUGCAGCUGGAACUGGCUUAGGUCGCCACUCGCA-GCAGCUGCACCA------------------ ......................(((((((((((((((((--((......))))))))...)))).)))))))......(((-(...))))....------------------ ( -30.70, z-score = -1.49, R) >anoGam1.chr3R 35999312 94 - 53272125 CGAACAGCAUGUCGUCGAAUCCGCACUCGAACUCGAACUCGACCCCAGCCUCGCCGCACCUGCUGUCGAACAGCUCGAGCGUCCACCUGCACCA------------------ ......(((.((.(.((.........(((....))).(((((.....((......))....((((.....))))))))))).).)).)))....------------------ ( -20.50, z-score = 0.40, R) >consensus CGCACAAAUCCGUUUCGCAUCUCGACUUGGCCACCAGCU__GCCAUGAUGCAGCUGGAACUGGCUUAGGUCGCCACUCGCA_GCAGCUGCACCA__________________ .......................((((((((((((((((..((......))))))))...)))).))))))......................................... (-16.96 = -17.17 + 0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:24 2011