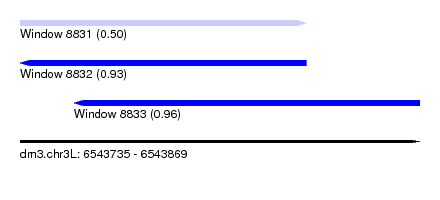
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,543,735 – 6,543,869 |
| Length | 134 |
| Max. P | 0.964978 |

| Location | 6,543,735 – 6,543,831 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 68.71 |
| Shannon entropy | 0.53684 |
| G+C content | 0.48717 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -12.96 |
| Energy contribution | -12.67 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
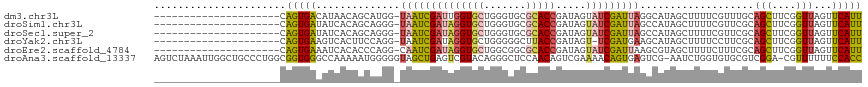
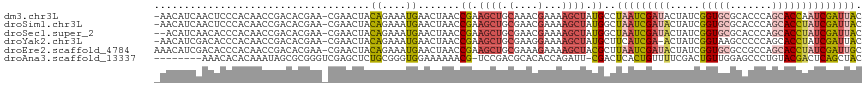
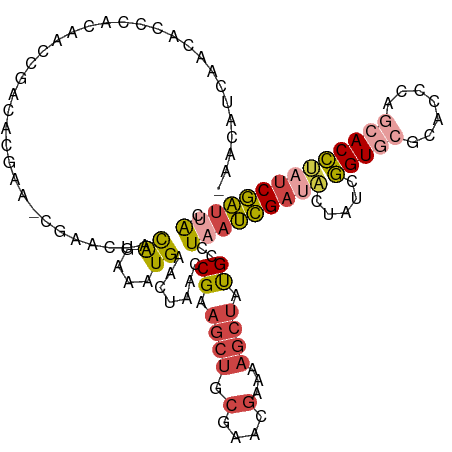
>dm3.chr3L 6543735 96 + 24543557 ---------------------CAGUGACAUAACAGCAUGG-UAAUCGAUUGGUGCUGGGUGCGCACCGAUAGUAUCGAUUAGGCAUAGCUUUUCGUUUGCAGCUUCGGUUAGUUCAUU ---------------------.(((((..(((((((....-((((((((((((((.......))))))).....))))))).(((.(((.....)))))).)))...))))..))))) ( -26.00, z-score = -0.69, R) >droSim1.chr3L 6014643 96 + 22553184 ---------------------CAGUGAUAUCACAGCAGGG-UAAUCGAUAGGUGCUGGGUGCGCACCGAUAGUAUCGAUUAGCCAUAGCUUUUCGUUCGCAGCUUCGGUUAGUUCAUU ---------------------..((((......(((.((.-((((((((((((((.......))))).....))))))))).))...)))......))))(((....)))........ ( -29.60, z-score = -2.15, R) >droSec1.super_2 6480215 96 + 7591821 ---------------------CAGUGAUAUCACAGCAGGG-UAAUCGAUAGGUGCUGGGUGCGCACCGAUAGUAUCGAUUAGCCAUAGCUUUUCGUUCGCAGCUUCGGUUAGUUCAUU ---------------------..((((......(((.((.-((((((((((((((.......))))).....))))))))).))...)))......))))(((....)))........ ( -29.60, z-score = -2.15, R) >droYak2.chr3L 7113516 95 + 24197627 ---------------------CAGUGAAGUCACUUCCAGG-UAAUCGAUAGGUGCUGGGGGCUUACCGAUAGU-UCGAUGAAGCAUAGCUUUUCCUUCGCAGCUUCGGUUAGUUCAUU ---------------------.((((....)))).((((.-..(((....))).))))(((((.(((((.(((-((((.(((((...)))))....))).))))))))).)))))... ( -30.10, z-score = -2.16, R) >droEre2.scaffold_4784 9214940 96 + 25762168 ---------------------CAGUGAAAUCACACCCAGG-CAAUCGAUAGGUGCUGGCGGCGCACCGAUAGUAUCGAUUAAGCGUAGCUUUUCUUUCGCAGCUUCGGUUAGUUCAUU ---------------------.((((((.....(((.(((-((((((((((((((.......))))).....))))))))..(((.((.....))..))).)))).)))...)))))) ( -28.20, z-score = -1.53, R) >droAna3.scaffold_13337 2900037 116 - 23293914 AGUCUAAAUUGGCUGCCCUGGCGGUGGGCCAAAAAUGGGGGUAGCUGAGUCGUACAGGGCUCCAACAGUCGAAAACAGUGAGUCG-AAUCUGGUGUGCGUCGGA-CGUUUUUUCCACC .((((...(..(((((((((((.....))).......))))))))..)(.((((((.((((.....)))).....(((.......-...))).)))))).))))-)............ ( -35.91, z-score = 0.21, R) >consensus _____________________CAGUGAAAUCACAGCAGGG_UAAUCGAUAGGUGCUGGGUGCGCACCGAUAGUAUCGAUUAGGCAUAGCUUUUCGUUCGCAGCUUCGGUUAGUUCAUU ......................(((((..............((((((((((((((.......))))).....)))))))))......((.........)).............))))) (-12.96 = -12.67 + -0.30)
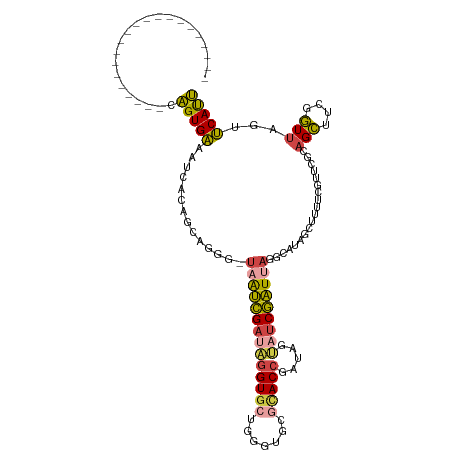
| Location | 6,543,735 – 6,543,831 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.71 |
| Shannon entropy | 0.53684 |
| G+C content | 0.48717 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -12.84 |
| Energy contribution | -13.82 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6543735 96 - 24543557 AAUGAACUAACCGAAGCUGCAAACGAAAAGCUAUGCCUAAUCGAUACUAUCGGUGCGCACCCAGCACCAAUCGAUUA-CCAUGCUGUUAUGUCACUG--------------------- ..(((..((((...((((..........))))..((.((((((((......(((((.......))))).))))))))-....)).))))..)))...--------------------- ( -20.30, z-score = -1.59, R) >droSim1.chr3L 6014643 96 - 22553184 AAUGAACUAACCGAAGCUGCGAACGAAAAGCUAUGGCUAAUCGAUACUAUCGGUGCGCACCCAGCACCUAUCGAUUA-CCCUGCUGUGAUAUCACUG--------------------- ............................(((...((.(((((((((.....(((((.......))))))))))))))-.)).)))(((....)))..--------------------- ( -24.60, z-score = -2.96, R) >droSec1.super_2 6480215 96 - 7591821 AAUGAACUAACCGAAGCUGCGAACGAAAAGCUAUGGCUAAUCGAUACUAUCGGUGCGCACCCAGCACCUAUCGAUUA-CCCUGCUGUGAUAUCACUG--------------------- ............................(((...((.(((((((((.....(((((.......))))))))))))))-.)).)))(((....)))..--------------------- ( -24.60, z-score = -2.96, R) >droYak2.chr3L 7113516 95 - 24197627 AAUGAACUAACCGAAGCUGCGAAGGAAAAGCUAUGCUUCAUCGA-ACUAUCGGUAAGCCCCCAGCACCUAUCGAUUA-CCUGGAAGUGACUUCACUG--------------------- ..........(((......((((((....(((..((((.(((((-....)))))))))....))).))).)))....-..))).((((....)))).--------------------- ( -19.50, z-score = -0.85, R) >droEre2.scaffold_4784 9214940 96 - 25762168 AAUGAACUAACCGAAGCUGCGAAAGAAAAGCUACGCUUAAUCGAUACUAUCGGUGCGCCGCCAGCACCUAUCGAUUG-CCUGGGUGUGAUUUCACUG--------------------- .........(((..((((.(....)...)))).((..(((((((((.....(((((.......))))))))))))))-..)))))(((....)))..--------------------- ( -28.00, z-score = -2.29, R) >droAna3.scaffold_13337 2900037 116 + 23293914 GGUGGAAAAAACG-UCCGACGCACACCAGAUU-CGACUCACUGUUUUCGACUGUUGGAGCCCUGUACGACUCAGCUACCCCCAUUUUUGGCCCACCGCCAGGGCAGCCAAUUUAGACU (((((......((-(.((..((.((.(((..(-(((..........))))))).))..))..)).)))......))))).......(((((...((....))...)))))........ ( -25.00, z-score = 1.04, R) >consensus AAUGAACUAACCGAAGCUGCGAACGAAAAGCUAUGCCUAAUCGAUACUAUCGGUGCGCACCCAGCACCUAUCGAUUA_CCCUGCUGUGAUAUCACUG_____________________ ..(((......((.((((.(....)...)))).))..(((((((((.....(((((.......))))))))))))))..............)))........................ (-12.84 = -13.82 + 0.98)
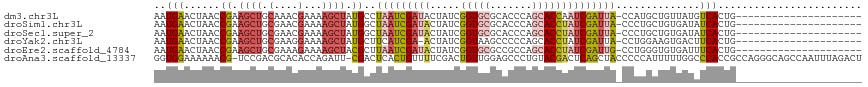
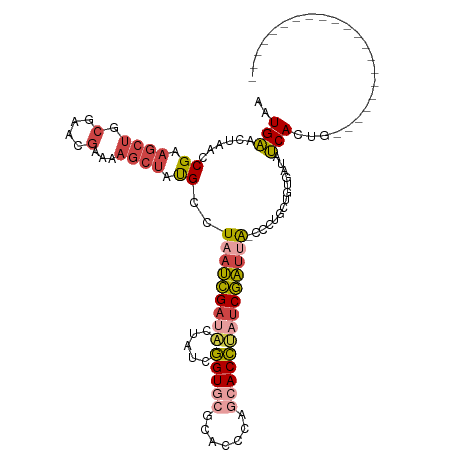
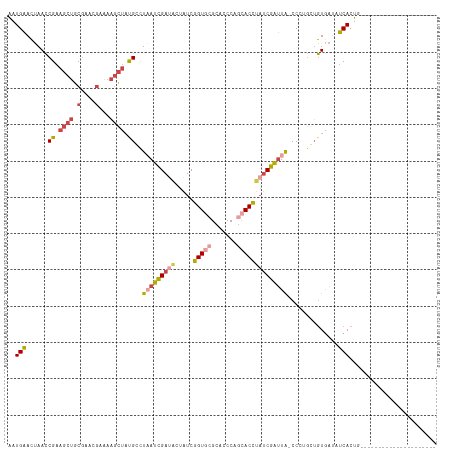
| Location | 6,543,753 – 6,543,869 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.10 |
| Shannon entropy | 0.45715 |
| G+C content | 0.48673 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -12.57 |
| Energy contribution | -13.55 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6543753 116 - 24543557 -AACAUCAACUCCCACAACCGACACGAA-CGAACUACAGAAAUGAACUAACCGAAGCUGCAAACGAAAAGCUAUGCCUAAUCGAUACUAUCGGUGCGCACCCAGCACCAAUCGAUUAC -.......................(((.-(((....((....))..........((((..........))))........)))........(((((.......)))))..)))..... ( -18.30, z-score = -1.96, R) >droSim1.chr3L 6014661 116 - 22553184 -AACAUCAACUCCCACAACCGACACGAA-CGAACUACAGAAAUGAACUAACCGAAGCUGCGAACGAAAAGCUAUGGCUAAUCGAUACUAUCGGUGCGCACCCAGCACCUAUCGAUUAC -...........................-.......((....))......(((.((((.(....)...)))).))).(((((((((.....(((((.......)))))))))))))). ( -23.70, z-score = -2.96, R) >droSec1.super_2 6480233 115 - 7591821 --ACAUCAACACCCACAACCGACACGAA-CGAACUACAGAAAUGAACUAACCGAAGCUGCGAACGAAAAGCUAUGGCUAAUCGAUACUAUCGGUGCGCACCCAGCACCUAUCGAUUAC --..........................-.......((....))......(((.((((.(....)...)))).))).(((((((((.....(((((.......)))))))))))))). ( -23.70, z-score = -3.15, R) >droYak2.chr3L 7113534 115 - 24197627 -AACAUCGACACCCACAACCGACACGAA-CGAACUACAGAAAUGAACUAACCGAAGCUGCGAAGGAAAAGCUAUGCUUCAUCGA-ACUAUCGGUAAGCCCCCAGCACCUAUCGAUUAC -...(((((........(((((......-(((....((....))........(((((.((.........))...))))).))).-....)))))..((.....)).....)))))... ( -22.30, z-score = -2.63, R) >droEre2.scaffold_4784 9214958 117 - 25762168 AAACAUCGACACCCACAACCGACACGAA-CGAACUACAGAAAUGAACUAACCGAAGCUGCGAAAGAAAAGCUACGCUUAAUCGAUACUAUCGGUGCGCCGCCAGCACCUAUCGAUUGC ....(((((...................-(((....((....))...(((.((.((((.(....)...)))).)).))).)))........(((((.......)))))..)))))... ( -25.10, z-score = -2.63, R) >droAna3.scaffold_13337 2900077 108 + 23293914 --------AAACACACAAAUAGCGCGGGUCGAGCUCUGCGGGUGGAAAAAACG-UCCGACGCACACCAGAUU-CGACUCACUGUUUUCGACUGUUGGAGCCCUGUACGACUCAGCUAC --------...........((((..((((((.((((((((..((((.......-)))).))))((.(((..(-(((..........))))))).))))))......)))))).)))). ( -29.80, z-score = -0.11, R) >consensus _AACAUCAACACCCACAACCGACACGAA_CGAACUACAGAAAUGAACUAACCGAAGCUGCGAACGAAAAGCUAUGCCUAAUCGAUACUAUCGGUGCGCACCCAGCACCUAUCGAUUAC ....................................((....)).......((.((((.(....)...)))).))..(((((((((.....(((((.......)))))))))))))). (-12.57 = -13.55 + 0.98)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:22 2011