
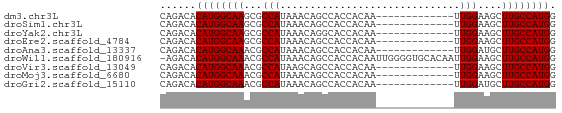
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,531,541 – 6,531,594 |
| Length | 53 |
| Max. P | 0.991214 |

| Location | 6,531,541 – 6,531,594 |
|---|---|
| Length | 53 |
| Sequences | 9 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 91.95 |
| Shannon entropy | 0.14860 |
| G+C content | 0.51972 |
| Mean single sequence MFE | -19.22 |
| Consensus MFE | -13.31 |
| Energy contribution | -13.31 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.991214 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 6531541 53 + 24543557 CAGACACAUGGCAAGCGCCAUAAACAGCCACCACAA-------------UUGGAAGCUUGCCAUGG ......((((((((((((........))..(((...-------------.)))..)))))))))). ( -21.10, z-score = -4.69, R) >droSim1.chr3L 6002622 53 + 22553184 CAGACACAUGGCAAGCGCCAUAAACAGCCACCACAA-------------UUGGAAGCUUGCCAUGG ......((((((((((((........))..(((...-------------.)))..)))))))))). ( -21.10, z-score = -4.69, R) >droYak2.chr3L 7101500 53 + 24197627 CAGACACAUGGCAAGCGCCAUAAACAGGCACCACAA-------------UUGGAAGCUUGCCAUGG ......(((((((((((((.......))).(((...-------------.)))..)))))))))). ( -25.50, z-score = -6.34, R) >droEre2.scaffold_4784 9203058 53 + 25762168 CAGACACAUGGCAAGCGCCAUAAACAGCCACCACAA-------------UUGGAAGCUUGCCAUGG ......((((((((((((........))..(((...-------------.)))..)))))))))). ( -21.10, z-score = -4.69, R) >droAna3.scaffold_13337 2858909 53 - 23293914 CAGACACAUGGCAAACGCCAUAAACAGCCACCACAA-------------UUGGAUGCUUGCCAUGG ......((((((((.(((((................-------------.))).)).)))))))). ( -15.23, z-score = -2.46, R) >droWil1.scaffold_180916 888225 65 + 2700594 -AGACACAUGGCAAACGCCAUAAACAGCCACCACAAUUGGGGUGCACAAUUGGAAGCUUGCCAUGG -.....((((((((...(((......(((.(((....)))))).......)))....)))))))). ( -22.32, z-score = -2.59, R) >droVir3.scaffold_13049 10886749 53 + 25233164 CAGACACAUGGCAAACGCCAUAAGCAGCCACCACAA-------------UUGGAAGCUUGCCAUGG ......((((((((..((.....)).((..(((...-------------.)))..)))))))))). ( -16.90, z-score = -2.85, R) >droMoj3.scaffold_6680 14825492 53 - 24764193 CAGACACAUGGCAAACGCCAUAAACAGCCACCACAA-------------UUGGAAGCUUGCCAUGG ......((((((((.(((........))..(((...-------------.)))..).)))))))). ( -14.50, z-score = -2.50, R) >droGri2.scaffold_15110 16138296 53 - 24565398 CAGACACAUGGCAAACGCCAUAAACAGCCACCACAA-------------UUGGAUGCUUGCCAUGG ......((((((((.(((((................-------------.))).)).)))))))). ( -15.23, z-score = -2.46, R) >consensus CAGACACAUGGCAAACGCCAUAAACAGCCACCACAA_____________UUGGAAGCUUGCCAUGG ......((((((((...(((..............................)))....)))))))). (-13.31 = -13.31 + -0.00)


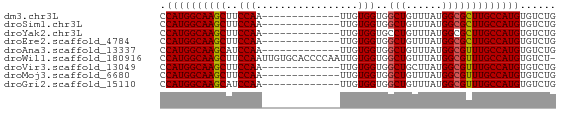
| Location | 6,531,541 – 6,531,594 |
|---|---|
| Length | 53 |
| Sequences | 9 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 91.95 |
| Shannon entropy | 0.14860 |
| G+C content | 0.51972 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -18.42 |
| Energy contribution | -18.29 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984096 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 6531541 53 - 24543557 CCAUGGCAAGCUUCCAA-------------UUGUGGUGGCUGUUUAUGGCGCUUGCCAUGUGUCUG .((((((((((..(((.-------------...)))..(((......)))))))))))))...... ( -21.60, z-score = -2.94, R) >droSim1.chr3L 6002622 53 - 22553184 CCAUGGCAAGCUUCCAA-------------UUGUGGUGGCUGUUUAUGGCGCUUGCCAUGUGUCUG .((((((((((..(((.-------------...)))..(((......)))))))))))))...... ( -21.60, z-score = -2.94, R) >droYak2.chr3L 7101500 53 - 24197627 CCAUGGCAAGCUUCCAA-------------UUGUGGUGCCUGUUUAUGGCGCUUGCCAUGUGUCUG .((((((((((..(((.-------------...))).(((.......)))))))))))))...... ( -23.60, z-score = -4.21, R) >droEre2.scaffold_4784 9203058 53 - 25762168 CCAUGGCAAGCUUCCAA-------------UUGUGGUGGCUGUUUAUGGCGCUUGCCAUGUGUCUG .((((((((((..(((.-------------...)))..(((......)))))))))))))...... ( -21.60, z-score = -2.94, R) >droAna3.scaffold_13337 2858909 53 + 23293914 CCAUGGCAAGCAUCCAA-------------UUGUGGUGGCUGUUUAUGGCGUUUGCCAUGUGUCUG .((((((((((..(((.-------------...)))..(((......)))))))))))))...... ( -19.70, z-score = -2.18, R) >droWil1.scaffold_180916 888225 65 - 2700594 CCAUGGCAAGCUUCCAAUUGUGCACCCCAAUUGUGGUGGCUGUUUAUGGCGUUUGCCAUGUGUCU- .((((((((((..(((.....((((((((....))).)).)))...))).)))))))))).....- ( -24.10, z-score = -2.30, R) >droVir3.scaffold_13049 10886749 53 - 25233164 CCAUGGCAAGCUUCCAA-------------UUGUGGUGGCUGCUUAUGGCGUUUGCCAUGUGUCUG .(((((((((((.(((.-------------...))).))))((.....))...)))))))...... ( -19.30, z-score = -2.00, R) >droMoj3.scaffold_6680 14825492 53 + 24764193 CCAUGGCAAGCUUCCAA-------------UUGUGGUGGCUGUUUAUGGCGUUUGCCAUGUGUCUG .((((((((((..(((.-------------...)))..(((......)))))))))))))...... ( -19.20, z-score = -2.42, R) >droGri2.scaffold_15110 16138296 53 + 24565398 CCAUGGCAAGCAUCCAA-------------UUGUGGUGGCUGUUUAUGGCGUUUGCCAUGUGUCUG .((((((((((..(((.-------------...)))..(((......)))))))))))))...... ( -19.70, z-score = -2.18, R) >consensus CCAUGGCAAGCUUCCAA_____________UUGUGGUGGCUGUUUAUGGCGUUUGCCAUGUGUCUG .((((((((((..(((.................)))..(((......)))))))))))))...... (-18.42 = -18.29 + -0.14)
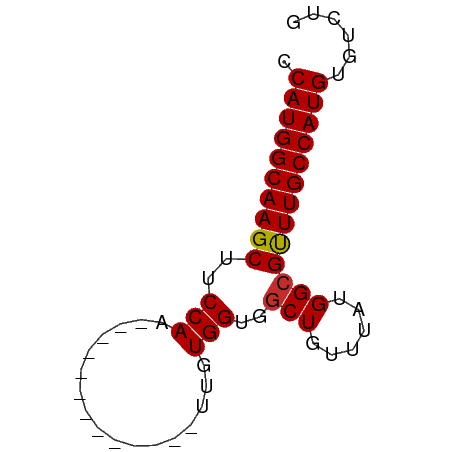
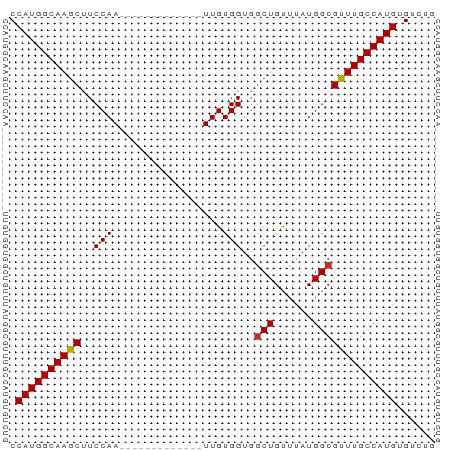
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:17 2011