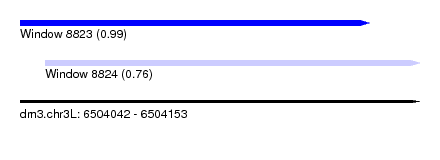
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,504,042 – 6,504,153 |
| Length | 111 |
| Max. P | 0.986951 |

| Location | 6,504,042 – 6,504,139 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.13 |
| Shannon entropy | 0.42389 |
| G+C content | 0.42464 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -17.68 |
| Energy contribution | -17.30 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.986951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
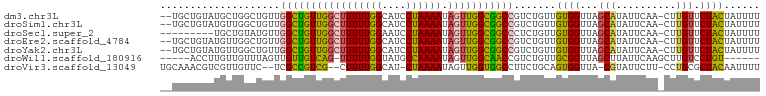
>dm3.chr3L 6504042 97 + 24543557 --UGCUGUAUGCUGGCUGUUGGCUGUUGGCUUUUUGGCAUCCUAAAAUAGUUGGCGGCCGUCUGUUGUGGUUAGCAUAUUCAA-CUUGUUCUACUAUUUU --((..((((((((((..((((((((..(((((((((....)))))).)))..)))))))......)..)))))))))).)).-................ ( -33.00, z-score = -3.48, R) >droSim1.chr3L 5975173 97 + 22553184 --UGCUGUAUGUUGGCUGUUGGCUGUUGGCUUUUUGGCAUCCUAAAAUAGUUGGCGGCCGUCUGUUGUGGUUAGCAUAUUCAA-CUUGUUCUACUAUUUU --((..((((((((((..((((((((..(((((((((....)))))).)))..)))))))......)..)))))))))).)).-................ ( -30.60, z-score = -3.05, R) >droSec1.super_2 6441205 90 + 7591821 ---------UGCUGUAUGUUGGCUGUUGGCUUUUUGGAAUCCUAAAAUAGUUGGCGGCCCUCUGUUGUGGUUAGCAUAUUCAA-CUUGUUCUACUAUUUU ---------((..(((((((((((((..(((((((((....)))))).)))..)))))((.(....).)).)))))))).)).-................ ( -23.60, z-score = -2.00, R) >droEre2.scaffold_4784 9174184 97 + 25762168 --UGCUGUAUGUUGGCUGUUGGCUGUUGGCUUUUUGGCAUCCUAAAAUAGUUGGCGGCCGUCUGUUGUGGUUAGCAUAUUCAA-CUUGUUCUACUAUUUU --((..((((((((((..((((((((..(((((((((....)))))).)))..)))))))......)..)))))))))).)).-................ ( -30.60, z-score = -3.05, R) >droYak2.chr3L 7072855 97 + 24197627 --UGCUGUAUGUUGGCUGUUGGCUGUUGGCUUUUUGGCAUCCUAAAAUAGUUGGCGGCCGUCUGUUGUGGUUAGCAUAUUCAA-CUUGUUCUACUAUUUU --((..((((((((((..((((((((..(((((((((....)))))).)))..)))))))......)..)))))))))).)).-................ ( -30.60, z-score = -3.05, R) >droWil1.scaffold_180916 1776252 88 - 2700594 -----ACCUUGUUGUUUAGUUGUUGUCAG-UUUUUGGUAUGCCAAAAUAGUUGGCAACCGUCUGUUGCGGUUAGCUUAUUCAAGCUUGUCCUGU------ -----........((.(((..((((((((-.((((((....))))))...))))))))...)))..))((..(((((....)))))...))...------ ( -22.60, z-score = -2.31, R) >droVir3.scaffold_13049 10862309 93 + 25233164 UGCAAACGUCGUUGUUC--UCGCCGUCG--CUUUUGGCAU-CUAAAAUAGUUGGUGGCCUUCUGCAGUGGUUA-CGUAUUCUU-CCUGCGCUACAAUUUU .(((((((....)))).--..((((..(--(....(((..-((((.....))))..)))....))..))))..-.........-..)))........... ( -17.00, z-score = 1.07, R) >consensus __UGCUGUAUGUUGGCUGUUGGCUGUUGGCUUUUUGGCAUCCUAAAAUAGUUGGCGGCCGUCUGUUGUGGUUAGCAUAUUCAA_CUUGUUCUACUAUUUU ....................(((((((((((((((((....)))))).))))))))))).......((((...(((..........))).))))...... (-17.68 = -17.30 + -0.38)
| Location | 6,504,049 – 6,504,153 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.32 |
| Shannon entropy | 0.32288 |
| G+C content | 0.40103 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -19.55 |
| Energy contribution | -19.19 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.761635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
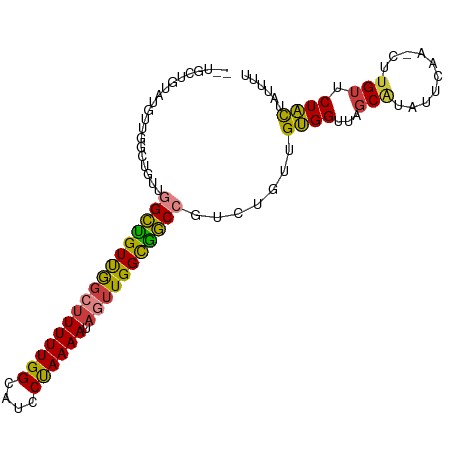
>dm3.chr3L 6504049 104 + 24543557 UGCUGGCUGUUGGCUGUUGGCUUUUUGGCAUCCUAAAAUAGUUGGCGGCCGUCUGUUGUGGUUAGCAUAUUCAA-CUUGUUCUACUAUUUUUAUACCCAUUUUGU (((((((..((((((((..(((((((((....)))))).)))..)))))))......)..))))))).......-.............................. ( -29.00, z-score = -1.84, R) >droSim1.chr3L 5975180 104 + 22553184 UGUUGGCUGUUGGCUGUUGGCUUUUUGGCAUCCUAAAAUAGUUGGCGGCCGUCUGUUGUGGUUAGCAUAUUCAA-CUUGUUCUACUAUUUUUAUACCCAUUUUGU (((((((..((((((((..(((((((((....)))))).)))..)))))))......)..))))))).......-.............................. ( -26.60, z-score = -1.34, R) >droSec1.super_2 6441212 97 + 7591821 -------UGUUGGCUGUUGGCUUUUUGGAAUCCUAAAAUAGUUGGCGGCCCUCUGUUGUGGUUAGCAUAUUCAA-CUUGUUCUACUAUUUUUAUACCCAUUUUGU -------.(..((((((..(((((((((....)))))).)))..))))))..)....(((((.((((.......-..))))..)))))................. ( -23.60, z-score = -1.64, R) >droEre2.scaffold_4784 9174191 104 + 25762168 UGUUGGCUGUUGGCUGUUGGCUUUUUGGCAUCCUAAAAUAGUUGGCGGCCGUCUGUUGUGGUUAGCAUAUUCAA-CUUGUUCUACUAUUUUUAUACCCAUUUUGU (((((((..((((((((..(((((((((....)))))).)))..)))))))......)..))))))).......-.............................. ( -26.60, z-score = -1.34, R) >droYak2.chr3L 7072862 104 + 24197627 UGUUGGCUGUUGGCUGUUGGCUUUUUGGCAUCCUAAAAUAGUUGGCGGCCGUCUGUUGUGGUUAGCAUAUUCAA-CUUGUUCUACUAUUUUUAUACCCAUUUUGU (((((((..((((((((..(((((((((....)))))).)))..)))))))......)..))))))).......-.............................. ( -26.60, z-score = -1.34, R) >droAna3.scaffold_13337 2832811 101 - 23293914 --UCUGUUGCUG-UUGUUGGCUUUUUGGCAUCCUAAAAUAGUUGGCGGCCGUCUGUUGUGGUUAGCAUAUUCAG-CUUGUUCUACUAUUUUUAAACCCAUUUUGU --......((.(-((((..(((((((((....)))))).)))..))))).)).....((((..(((.......)-))....)))).................... ( -19.60, z-score = 0.31, R) >droWil1.scaffold_180916 1776256 84 - 2700594 UGUUGUUUAGUUGUUGUCAG-UUUUUGGUAUGCCAAAAUAGUUGGCAACCGUCUGUUGCGGUUAGCUUAUUCAAGCUUGUCCUGU-------------------- ....((.(((..((((((((-.((((((....))))))...))))))))...)))..))((..(((((....)))))...))...-------------------- ( -22.60, z-score = -2.77, R) >consensus UGUUGGCUGUUGGCUGUUGGCUUUUUGGCAUCCUAAAAUAGUUGGCGGCCGUCUGUUGUGGUUAGCAUAUUCAA_CUUGUUCUACUAUUUUUAUACCCAUUUUGU ...........((((((..(((((((((....)))))).)))..)))))).......((((...(((..........))).)))).................... (-19.55 = -19.19 + -0.36)


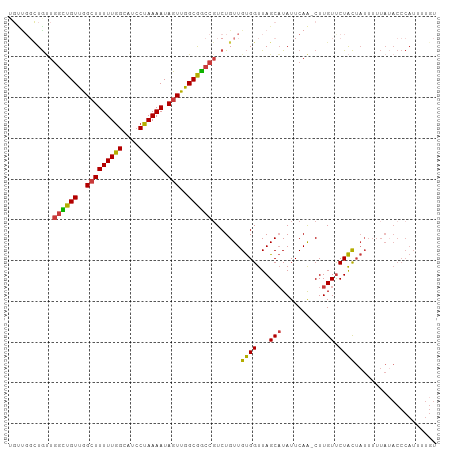
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:15 2011