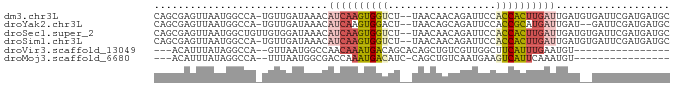
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,501,004 – 6,501,087 |
| Length | 83 |
| Max. P | 0.757854 |

| Location | 6,501,004 – 6,501,087 |
|---|---|
| Length | 83 |
| Sequences | 6 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 62.26 |
| Shannon entropy | 0.64668 |
| G+C content | 0.40974 |
| Mean single sequence MFE | -16.50 |
| Consensus MFE | -7.96 |
| Energy contribution | -7.69 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.60 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.649007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
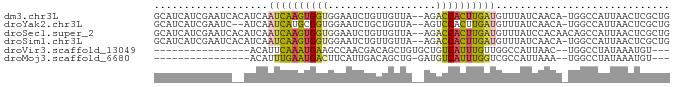
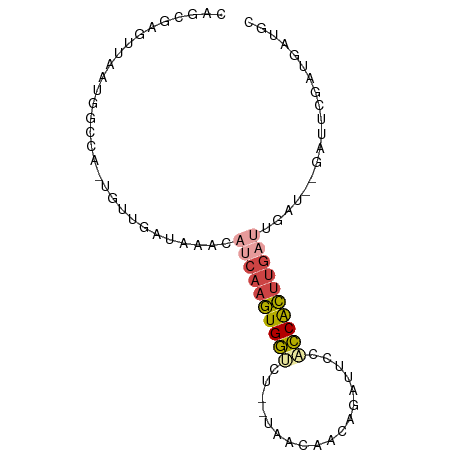
>dm3.chr3L 6501004 83 + 24543557 GCAUCAUCGAAUCACAUCAAUCAAGUGGUGGAAUCUGUUGUUA--AGACCACUUGAUGUUUAUCAACA-UGGCCAUUAACUCGCUG ((......((.((.(((((......))))))).))....((((--(..(((.(((((....)))))..-)))...)))))..)).. ( -15.50, z-score = 0.14, R) >droYak2.chr3L 7069788 81 + 24197627 GCAUCAUCGAAUC--AUCAAUCAUGCGGUGGAAUCUGCUGUUA--AGUCCACUUGAUGUUUAUCAACA-UGGCCAUUAACUCGCUG ((...........--.........((((......)))).((((--(..(((.(((((....)))))..-)))...)))))..)).. ( -13.60, z-score = 0.69, R) >droSec1.super_2 6438201 84 + 7591821 GCAUCAUCGAAUCACAUCAAUCAAGUGGUGGAAUCUGUUGUUA--AGACCACUUGAUGUUUAUCCACAACAGCCAUUAACUCGCUG ...................((((((((((..(((.....))).--..))))))))))............((((.........)))) ( -15.60, z-score = -0.30, R) >droSim1.chr3L 5972158 83 + 22553184 GCAUCAUCGAAUCACAUCAAUCAAGUGGUGGAAUCUGUUGUUA--AGACCACUUGAUGUUUAUCAACA-UGGCCAUUAACUCGCUG ((......((.((.(((((......))))))).))....((((--(..(((.(((((....)))))..-)))...)))))..)).. ( -15.50, z-score = 0.14, R) >droVir3.scaffold_13049 10859257 65 + 25233164 ----------------ACAUUCAAAUGAAGCCAACGACAGCUGUGCUGUCAUUUGUUGGCCAUUAAC--UGGCCUAUAAAUGU--- ----------------...(((....)))......((((((...))))))((((((.(((((.....--))))).))))))..--- ( -20.10, z-score = -2.83, R) >droMoj3.scaffold_6680 14798921 64 - 24764193 ----------------ACAUUUGAAUGACUUCAUUGACAGCUG-GAUGUCAUUUGGUCGCCAUUAAA--UGGCCUAUAAAUGU--- ----------------(((((((...((((....(((((....-..)))))...))))(((((...)--))))...)))))))--- ( -18.70, z-score = -2.76, R) >consensus GCAUCAUCGAAUC__AUCAAUCAAGUGGUGGAAUCUGCUGUUA__AGACCACUUGAUGUUUAUCAACA_UGGCCAUUAACUCGCUG ...................((((((((((..................))))))))))............................. ( -7.96 = -7.69 + -0.27)
| Location | 6,501,004 – 6,501,087 |
|---|---|
| Length | 83 |
| Sequences | 6 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 62.26 |
| Shannon entropy | 0.64668 |
| G+C content | 0.40974 |
| Mean single sequence MFE | -19.88 |
| Consensus MFE | -6.99 |
| Energy contribution | -7.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.757854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
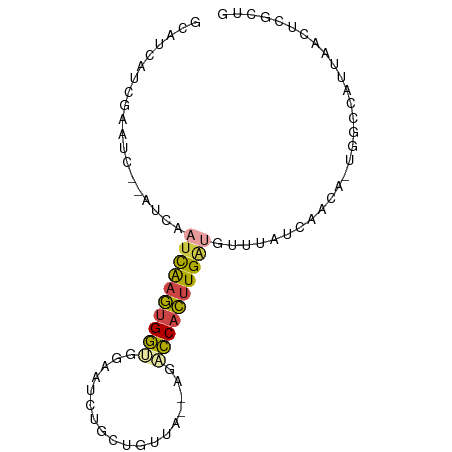
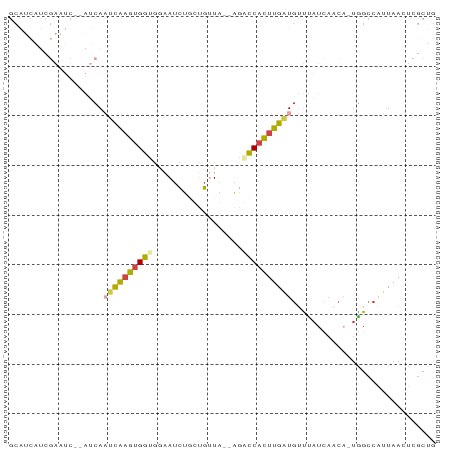
>dm3.chr3L 6501004 83 - 24543557 CAGCGAGUUAAUGGCCA-UGUUGAUAAACAUCAAGUGGUCU--UAACAACAGAUUCCACCACUUGAUUGAUGUGAUUCGAUGAUGC ((.(((((((.......-((((....))))(((((((((..--..............)))))))))......))))))).)).... ( -19.29, z-score = -0.75, R) >droYak2.chr3L 7069788 81 - 24197627 CAGCGAGUUAAUGGCCA-UGUUGAUAAACAUCAAGUGGACU--UAACAGCAGAUUCCACCGCAUGAUUGAU--GAUUCGAUGAUGC ..((..(((((...(((-(.(((((....)))))))))..)--)))).))..........((((.(((((.--...))))).)))) ( -21.50, z-score = -1.77, R) >droSec1.super_2 6438201 84 - 7591821 CAGCGAGUUAAUGGCUGUUGUGGAUAAACAUCAAGUGGUCU--UAACAACAGAUUCCACCACUUGAUUGAUGUGAUUCGAUGAUGC ((((.(.....).))))(..(.((...((((((((((((..--..............))))))....))))))...)).)..)... ( -19.59, z-score = -0.40, R) >droSim1.chr3L 5972158 83 - 22553184 CAGCGAGUUAAUGGCCA-UGUUGAUAAACAUCAAGUGGUCU--UAACAACAGAUUCCACCACUUGAUUGAUGUGAUUCGAUGAUGC ((.(((((((.......-((((....))))(((((((((..--..............)))))))))......))))))).)).... ( -19.29, z-score = -0.75, R) >droVir3.scaffold_13049 10859257 65 - 25233164 ---ACAUUUAUAGGCCA--GUUAAUGGCCAACAAAUGACAGCACAGCUGUCGUUGGCUUCAUUUGAAUGU---------------- ---(((((....(((((--.....)))))..(((((((.(((.((((....)))))))))))))))))))---------------- ( -24.90, z-score = -4.36, R) >droMoj3.scaffold_6680 14798921 64 + 24764193 ---ACAUUUAUAGGCCA--UUUAAUGGCGACCAAAUGACAUC-CAGCUGUCAAUGAAGUCAUUCAAAUGU---------------- ---((((((....((((--(...)))))(((..(.(((((..-....))))).)...)))....))))))---------------- ( -14.70, z-score = -1.89, R) >consensus CAGCGAGUUAAUGGCCA_UGUUGAUAAACAUCAAGUGGUCU__UAACAACAGAUUCCACCACUUGAUUGAU__GAUUCGAUGAUGC .............................((((((((((..................))))))))))................... ( -6.99 = -7.30 + 0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:12 2011