| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,437,030 – 6,437,132 |
| Length | 102 |
| Max. P | 0.934584 |

| Location | 6,437,030 – 6,437,132 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Shannon entropy | 0.36285 |
| G+C content | 0.37689 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -14.13 |
| Energy contribution | -14.26 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
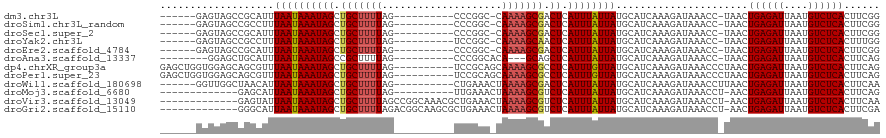
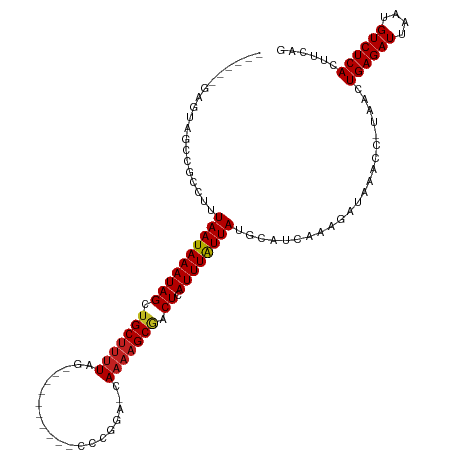
>dm3.chr3L 6437030 102 + 24543557 ------GAGUAGCCGCAUUUAAUAAAUAGCUGCUUUUAG----------CCCGGC-CAAAAGCGACUCAUUUAUUAUGCAUCAAAGAUAAACC-UAACUGAGAUUAAUGUCUCACUUCGG ------........(((..((((((((((.(((((((.(----------(...))-.))))))).)).)))))))))))............((-.((.((((((....)))))).)).)) ( -23.40, z-score = -2.15, R) >droSim1.chr3L_random 310033 102 + 1049610 ------GAGUAGCCGCCUUUAAUAAAUAGCUGCUUUUAG----------CCCGGC-CAAAAGCGACUCAUUUAUUAUGCAUCAAAGAUAAACC-UAACUGAGAUUAAUGUCUCACUUCGG ------........((...((((((((((.(((((((.(----------(...))-.))))))).)).)))))))).))............((-.((.((((((....)))))).)).)) ( -22.40, z-score = -1.78, R) >droSec1.super_2 6374735 102 + 7591821 ------GAGUAGCCGCAUUUAAUAAAUAGCUGCUUUUAG----------CCCGGC-CAAAAGCGACUCAUUUAUUAUGCAUCAAAGAUAAACC-UAACUGAGAUUAAUGUCUCACUUCGG ------........(((..((((((((((.(((((((.(----------(...))-.))))))).)).)))))))))))............((-.((.((((((....)))))).)).)) ( -23.40, z-score = -2.15, R) >droYak2.chr3L 7002318 102 + 24197627 ------GAGUAGCCGCCUUUAAUAAAUAGCUGCUUUUAG----------UCCGGC-CAAAAGCAACUCAUUUAUUAUGCAUCAAAGAUAAACC-UAACUGAGAUUAAUGUCUCACUUUGG ------........((...((((((((((.(((((((.(----------.....)-.))))))).)).)))))))).))..(((((.......-....((((((....))))))))))). ( -22.00, z-score = -1.96, R) >droEre2.scaffold_4784 9102348 102 + 25762168 ------GAGUAGCCGCAUUUAAUAAAUAGCUGCUUUUAG----------CCCGGC-CAAAAGCGACUCAUUUAUUAUGCAUCAAAGAUAAACC-UAACUGAGAUUAAUGUCUCACUUCGG ------........(((..((((((((((.(((((((.(----------(...))-.))))))).)).)))))))))))............((-.((.((((((....)))))).)).)) ( -23.40, z-score = -2.15, R) >droAna3.scaffold_13337 2765761 98 - 23293914 --------GGAGCUGCAUUUAAUAAAUAGCCGCUUUUAG----------CCCGGCACA---GCAGCUCAUUUAUUAUGCAUCAAAGAUAAACC-UAACUGAGAUUAAUGUCUCACUUCAG --------.((((((((((.....))).((((.......----------..))))...---))))))).((((((.(......).))))))..-....((((((....))))))...... ( -22.80, z-score = -1.91, R) >dp4.chrXR_group3a 471374 110 + 1468910 GAGCUGGUGGAGCAGCGUUUAAUAAAUAGCUGCUUUUAG----------UCCGCAGCAAAAGCGCCUCAUUUGUUAUGCAUCAAAGAUAAACCCUAACUGAGAUUAAUGUCUCACUUCAG ((((((......))))((.((((((((((.(((((((.(----------(.....))))))))).)).)))))))).)).))................((((((....))))))...... ( -25.10, z-score = -0.34, R) >droPer1.super_23 480463 110 + 1662726 GAGCUGGUGGAGCAGCGUUUAAUAAAUAGCUGCUUUUAG----------UCCGCAGCAAAAGCGCCUCAUUUGUUAUGCAUCAAAGAUAAACCCUAACUGAGAUUAAUGUCUCACUUCAG ((((((......))))((.((((((((((.(((((((.(----------(.....))))))))).)).)))))))).)).))................((((((....))))))...... ( -25.10, z-score = -0.34, R) >droWil1.scaffold_180698 6422668 104 - 11422946 ------GGUUGGCUAACAUUAAUAAAUAGCUGCUUUUAG----------CUGAAACUAAAAGCGACUCAUUUAUUAUGCAUCAAAGAUAAACCUUAACUGAGAUUAAUGUCUCACUUCAA ------((((..((.....((((((((((.(((((((((----------......))))))))).)).))))))))........))...)))).....((((((....))))))...... ( -24.32, z-score = -3.35, R) >droMoj3.scaffold_6680 10042768 96 + 24764193 -------------GAGCAUUAAUAAAUAGCUGCUUUUAG----------UUGAAACUAAAAGCGUCUCAUUUAUUAUGCAUCAAAGAUAAACCU-AACUGAGAUUAAUGUCUCACUUCAG -------------((((((.(((((((((.(((((((((----------(....)))))))))).)).))))))))))).))............-...((((((....))))))...... ( -26.80, z-score = -4.75, R) >droVir3.scaffold_13049 14463164 106 - 25233164 -------------GAGUAUUAAUAAAUAGCUGCUUUUAGCCGGCAAACGCUGAAACUAAAAGCGUCUCAUUUAUUAUGCAUCAAAGAUAAACCU-AACUGAGAUUAAUGUCUCACUUCAA -------------((((((.(((((((((.(((((((((.((((....))))...))))))))).)).))))))))))).))............-...((((((....))))))...... ( -28.00, z-score = -4.16, R) >droGri2.scaffold_15110 8525132 106 + 24565398 -------------GGGCAUUAAUAAAUAGCUGCUUUUAGACGGCAAGCGCUGAAACUAAAAGCGUCUCAUUUAUUAUGCAUCAAAGAUAAACCU-AACUGAGAUUAAUGUCUCACUUCGA -------------..((((.(((((((((.(((((((((.((((....))))...))))))))).)).)))))))))))...............-...((((((....))))))...... ( -29.30, z-score = -3.52, R) >consensus ______GAGUAGCCGCCUUUAAUAAAUAGCUGCUUUUAG__________CCCGGA_CAAAAGCGACUCAUUUAUUAUGCAUCAAAGAUAAACC_UAACUGAGAUUAAUGUCUCACUUCAG ...................((((((((((.(((((((....................))))))).)).))))))))......................((((((....))))))...... (-14.13 = -14.26 + 0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:06 2011