| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,387,110 – 6,387,208 |
| Length | 98 |
| Max. P | 0.577779 |

| Location | 6,387,110 – 6,387,208 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.07 |
| Shannon entropy | 0.39695 |
| G+C content | 0.42657 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -12.87 |
| Energy contribution | -12.64 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6387110 98 + 24543557 AGGAAUUGGAAAUAAUAAGGCGUAACUAAAUGGGC-GCUUUAGUCUAUAAUCAUGACACAAUUUCCGCCGGAGUCUCGGGCUCACUCCAACUGCCAACU---- .(((...((((((..((((((((..........))-))))))(((.........)))...))))))(((.(.....).)))....)))...........---- ( -25.70, z-score = -1.62, R) >droSim1.chr3L 5885999 98 + 22553184 AGGAAUUGGAAAUAAUAAGGCGUAACUAAAUGGGC-GCUUUAGUCUAUAAUCAUGACACAAUUUCCGCCGUAGUCUCGGGCUCACUCCAACUGCCAACU---- .(((...((((((..((((((((..........))-))))))(((.........)))...))))))(((.........)))....)))...........---- ( -23.00, z-score = -1.02, R) >droSec1.super_2 6351236 91 + 7591821 AGGAAUUGGAAAUAAUAAGGCGUAACUAAAUGGGC-GCUUUAGUCUAUAAUCAUGACACAAUUUCCGCCGGAGUCUCGGGCUCACUCCAACU----------- .(((...((((((..((((((((..........))-))))))(((.........)))...))))))(((.(.....).)))....)))....----------- ( -25.70, z-score = -2.41, R) >droYak2.chr3L 6978113 86 + 24197627 AGGAAUUGGAAAUAAUAAGGCGUAACUAAAUGGGC-GCUUUAGUCUAUAAUCAUGACACAAUUUCCGCCGGAGUCUCGGGCUCAACU---------------- .......((((((..((((((((..........))-))))))(((.........)))...))))))(((.(.....).)))......---------------- ( -22.30, z-score = -1.71, R) >droEre2.scaffold_4784 9078975 98 + 25762168 AGGAAUUGGAAAUAAUAAGGCGUAACUAAAUGGGC-GCUUUAGUCUAUAAUCAUGACACAAUUUCCGCCGGAGUCUCGGGCUCACUCCAACUGCCAACU---- .(((...((((((..((((((((..........))-))))))(((.........)))...))))))(((.(.....).)))....)))...........---- ( -25.70, z-score = -1.62, R) >droAna3.scaffold_13337 2741131 89 - 23293914 AGGAAUUGGAAAUAAUAAGGCGUAACUAAAUGGGC-GCUUUAGUCUAUAAUCAUGACACAAUUUCCGCCGGAGUC--GGGCUCACUCGAACU----------- .((....((((((..((((((((..........))-))))))(((.........)))...)))))).))....((--(((....)))))...----------- ( -21.90, z-score = -1.23, R) >droWil1.scaffold_180698 5027624 98 + 11422946 AAAUAAGGGAAAUAAUAAGGCGUAACUAAAUAGGCCGCUUUAGUCUAUAAUCAUGACACAAUUUCCGCCACA-GC----CACCAGAUGAAACAUAAGCGAACA .......((((((..(((((((...((....))..)))))))(((.........)))...))))))......-..----........................ ( -15.40, z-score = -0.39, R) >droVir3.scaffold_13049 10746424 87 + 25233164 AGCUAAAGGAAAUAAUAAGGCGUAACUAAAUAGGC-GCUUUAGUCUAUAAUCAUGACACAAUUUCCGCCAGCCAC----AGCUCUCCGAAUC----------- .(((...((((((..((((((((..........))-))))))(((.........)))...))))))...)))...----.............----------- ( -19.40, z-score = -2.01, R) >droGri2.scaffold_15110 16015777 84 - 24565398 AGCCAAAGGAAAUAAUAAGGCGUAACUAAAUAGGC-GCUUUAGUCUAUAAUCAUGACACAAUUUCCGCCAG--AC----AGCCA--AGAAUUG---------- .......((((((..((((((((..........))-))))))(((.........)))...))))))((...--..----.))..--.......---------- ( -16.50, z-score = -1.56, R) >consensus AGGAAUUGGAAAUAAUAAGGCGUAACUAAAUGGGC_GCUUUAGUCUAUAAUCAUGACACAAUUUCCGCCGGAGUCUCGGGCUCACUCCAACU___________ .......((((((..((((((....((....))...))))))(((.........)))...))))))..................................... (-12.87 = -12.64 + -0.22)

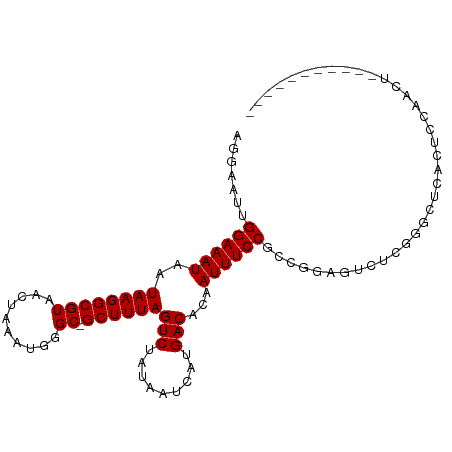

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:03 2011