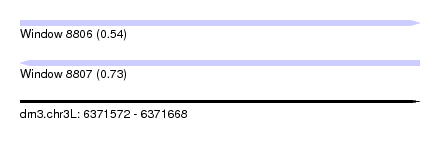
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,371,572 – 6,371,668 |
| Length | 96 |
| Max. P | 0.726463 |

| Location | 6,371,572 – 6,371,668 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 84.43 |
| Shannon entropy | 0.26126 |
| G+C content | 0.51042 |
| Mean single sequence MFE | -25.74 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.30 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.536677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
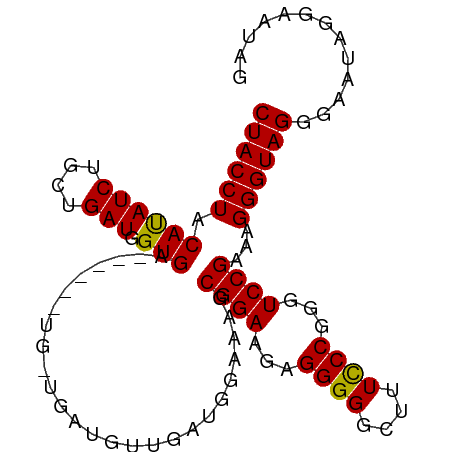
>dm3.chr3L 6371572 96 + 24543557 CUACCUACAUAUCUGCUGAUGGUGAGAU--GUGGUGAUGUUGAUGGAAAGCGGAAGAGGGGGCUUUCCCGGGUCCGAAAGGGUAGGGAAUAGGAAUAG ((((((.(((((((((.....)).))))--))).................((((...((((....))))...))))...))))))............. ( -27.40, z-score = -1.36, R) >droSim1.chr3L 5870528 80 + 22553184 CUACCUACAUAUCUGCUGAUGGUGA------UG------------GAAAGCGGAAGAGGGGGCUUUCCCGGGUCCGAAAGGGUAGGGAAUAGGAAUAG ...((((....((((((........------..------------...))))))..........(((((...(((....)))..)))))))))..... ( -23.32, z-score = -0.85, R) >droSec1.super_2 6335925 92 + 7591821 CUACCUACAUAUCUGCUGAUGGUGA------UGACGAUGGUGAUGGAAAGCGGAAGAGGGGGCUUUCCCGGGUCCGAAAGGGUAGGGAAUAGGAAUAG ...((((....((((((.....(.(------(.((....)).)).)..))))))..........(((((...(((....)))..)))))))))..... ( -25.90, z-score = -1.04, R) >droEre2.scaffold_4784 9062357 97 + 25762168 CUACCUACACAUCUGCUGAUGGUGAUGGUGAAGUUGAUGUUGAUGAAAAGCGGAAGAGGGG-CUUUUCCGGCUCCGAAAGGGUAGGGAAUUGGAAUAG ...((((((((((.(((..............))).)))))........((((((((((...-))))))).)))((....)))))))............ ( -26.34, z-score = -1.31, R) >consensus CUACCUACAUAUCUGCUGAUGGUGA______UG_UGAUGUUGAUGGAAAGCGGAAGAGGGGGCUUUCCCGGGUCCGAAAGGGUAGGGAAUAGGAAUAG ((((((.((((((....))).)))..........................((((...((((....))))...))))...))))))............. (-21.67 = -21.30 + -0.37)


| Location | 6,371,572 – 6,371,668 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 84.43 |
| Shannon entropy | 0.26126 |
| G+C content | 0.51042 |
| Mean single sequence MFE | -17.00 |
| Consensus MFE | -13.12 |
| Energy contribution | -13.38 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
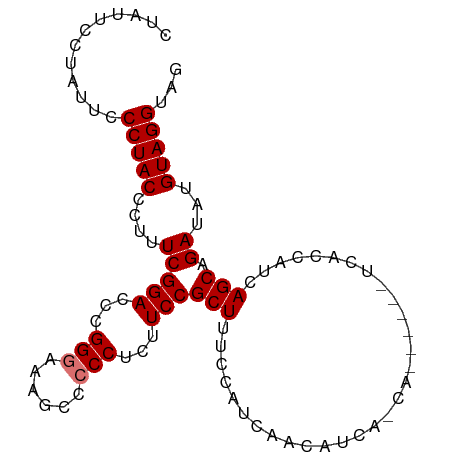
>dm3.chr3L 6371572 96 - 24543557 CUAUUCCUAUUCCCUACCCUUUCGGACCCGGGAAAGCCCCCUCUUCCGCUUUCCAUCAACAUCACCAC--AUCUCACCAUCAGCAGAUAUGUAGGUAG .............(((((....((....))(((((((..........)))))))............((--((.((..........)).)))).))))) ( -17.00, z-score = -1.75, R) >droSim1.chr3L 5870528 80 - 22553184 CUAUUCCUAUUCCCUACCCUUUCGGACCCGGGAAAGCCCCCUCUUCCGCUUUC------------CA------UCACCAUCAGCAGAUAUGUAGGUAG .............(((((.....((.....(((((((..........))))))------------).------...))....(((....))).))))) ( -17.10, z-score = -1.58, R) >droSec1.super_2 6335925 92 - 7591821 CUAUUCCUAUUCCCUACCCUUUCGGACCCGGGAAAGCCCCCUCUUCCGCUUUCCAUCACCAUCGUCA------UCACCAUCAGCAGAUAUGUAGGUAG .............(((((.....((.....(((((((..........)))))))....)).......------.........(((....))).))))) ( -17.10, z-score = -1.45, R) >droEre2.scaffold_4784 9062357 97 - 25762168 CUAUUCCAAUUCCCUACCCUUUCGGAGCCGGAAAAG-CCCCUCUUCCGCUUUUCAUCAACAUCAACUUCACCAUCACCAUCAGCAGAUGUGUAGGUAG .............(((((....((....))((((((-(.........))))))).....................((((((....)))).)).))))) ( -16.80, z-score = -1.85, R) >consensus CUAUUCCUAUUCCCUACCCUUUCGGACCCGGGAAAGCCCCCUCUUCCGCUUUCCAUCAACAUCA_CA______UCACCAUCAGCAGAUAUGUAGGUAG .............(((((....((((...(((......)))...))))..................................(((....))).))))) (-13.12 = -13.38 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:07:01 2011