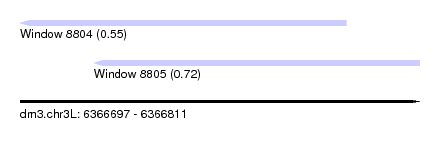
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,366,697 – 6,366,811 |
| Length | 114 |
| Max. P | 0.723305 |

| Location | 6,366,697 – 6,366,790 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 75.51 |
| Shannon entropy | 0.45078 |
| G+C content | 0.61963 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -17.90 |
| Energy contribution | -20.22 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
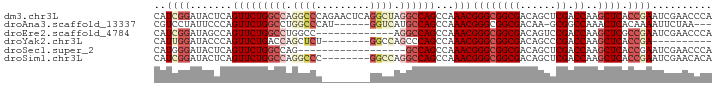
>dm3.chr3L 6366697 93 - 24543557 CAUCGGAUACUCAGUUCUGGCCAGGCCCAGAACUCAGGCUAGGCCAGCCAAACGGGCGGCGACAGCUCGACCAAGCUCACCGAAUCGAACCCA ..((((.......(((((((((.((((.........)))).))))))...)))(((((((((....))).))..)))).)))).......... ( -32.00, z-score = -1.69, R) >droAna3.scaffold_13337 2719363 83 + 23293914 CGUCCUAUUCCCAGUUCUGGCCUGGCCCAU------GGUCAUGCCAGCCAAACGGGCGGCGACAA-GCGGCCAAACUCACAAAAUUCUAA--- ............((((.((((((((((...------)))))((((.(((.....)))))))....-..))))))))).............--- ( -24.90, z-score = -0.77, R) >droEre2.scaffold_4784 9057574 80 - 25762168 CAUCGGAUAGCCAGUUCUGGCCUGGCC-------------AGGCCAGCCAAACGGGCGGCGACAGUCCGACCAAGCUCGCCGAAUCGAACCCA ..((((..(((.....(((((((....-------------))))))).....((((((....).))))).....)))..)))).......... ( -29.80, z-score = -1.15, R) >droYak2.chr3L 6956280 75 - 24197627 CAUUGGAUACCCAGUUCUGACCAGCUCU--------GGCCAGCCCAGCCAAACGGGCGGCGACAGCCCGACCAAGCUCACCGA---------- .(((((....))))).......((((.(--------((.(.((((........))))(((....))).).)))))))......---------- ( -22.40, z-score = -0.54, R) >droSec1.super_2 6331124 75 - 7591821 CAUGGGAUACUCAGUUCUGGCCAG------------------GCCAGCCAAACGGGCGGCGACAGCUCGACCAAGCUCACCGAAUCGAACCCA ..((((....((.((((.((....------------------(((.(((.....))))))...((((......))))..)))))).)).)))) ( -20.50, z-score = 0.18, R) >droSim1.chr3L 5865136 85 - 22553184 CAUCGGAUACUCAGUUCUGGCCAGGCCC--------GGCCAGGCCAGCCAAACGGGCGGCGACAGCUCGACCAAGCUCACCGAAUCGAACACA ..((((.......(((((((((......--------))))))(((.(((.....)))))))))((((......))))..)))).......... ( -29.90, z-score = -1.59, R) >consensus CAUCGGAUACCCAGUUCUGGCCAGGCCC________GGCCAGGCCAGCCAAACGGGCGGCGACAGCUCGACCAAGCUCACCGAAUCGAACCCA ..((((.......(((((((((.((((.........)))).))))))...)))((((((((......)).))..)))).)))).......... (-17.90 = -20.22 + 2.32)
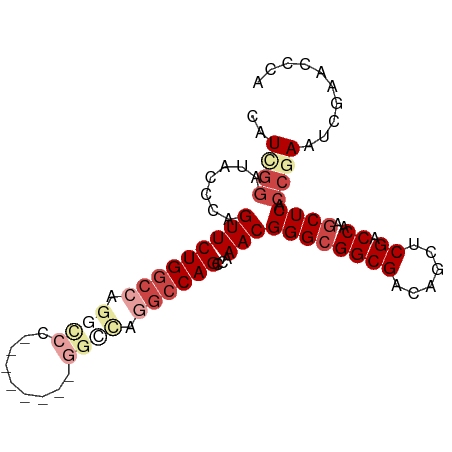
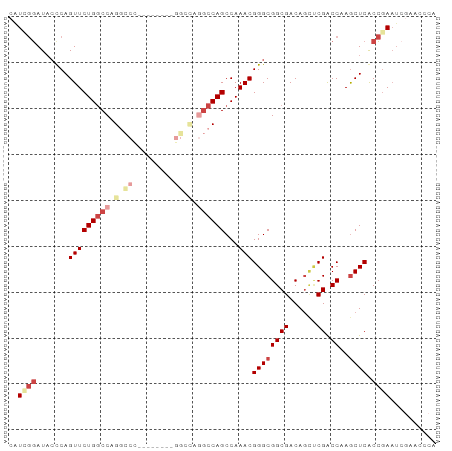
| Location | 6,366,718 – 6,366,811 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 72.74 |
| Shannon entropy | 0.41762 |
| G+C content | 0.65295 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -19.67 |
| Energy contribution | -22.80 |
| Covariance contribution | 3.13 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6366718 93 - 24543557 CCAUUCCCUAGCCCAAUCCCCCAUCGGAUACUCAGUUCUGGCCAGGCCCAGAACUCAGGCUAGGCCAGCCAAACGGGCGGCGACAGCUCGACC ......(((((((..((((......))))....((((((((......))))))))..))))))).........((((((....).)))))... ( -33.60, z-score = -2.11, R) >droAna3.scaffold_13337 2719381 73 + 23293914 -------------CAAUUCCCCGUCCUAUUCCCAGUUCUGGCCUGGCCCA------UGGUCAUGCCAGCCAAACGGGCGGCGACAAGCGGCC- -------------....((.((((((........((((((((.(((((..------.))))).)))))...))))))))).)).........- ( -23.30, z-score = 0.07, R) >droEre2.scaffold_4784 9057595 80 - 25762168 CCAUUCCCUAACCCAAUCCCCCAUCGGAUAGCCAGUUCUGGCCU-------------GGCCAGGCCAGCCAAACGGGCGGCGACAGUCCGACC .......................((((((.(((.....((((((-------------....))))))(((.....))))))....)))))).. ( -28.00, z-score = -1.69, R) >droSim1.chr3L 5865157 79 - 22553184 ------CCUAGCCCAAUCCCCCAUCGGAUACUCAGUUCUGGCCAGGCCC--------GGCCAGGCCAGCCAAACGGGCGGCGACAGCUCGACC ------....((((.((((......)))).....(((((((((.(((..--------.))).))))))...)))))))(((((....))).)) ( -30.10, z-score = -1.32, R) >consensus ______CCUAGCCCAAUCCCCCAUCGGAUACCCAGUUCUGGCCAGGCCC________GGCCAGGCCAGCCAAACGGGCGGCGACAGCCCGACC ..........((((.((((......)))).....((((((((((((((.........)))))))))))...)))))))(((....)))..... (-19.67 = -22.80 + 3.13)

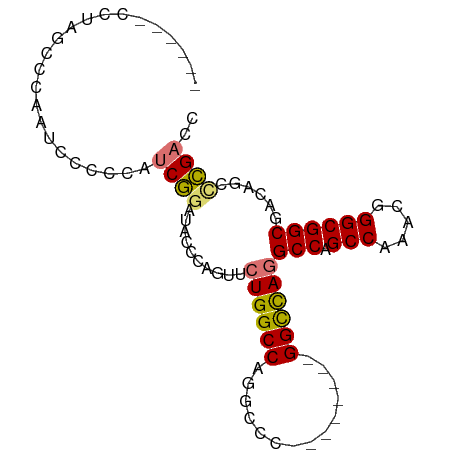
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:59 2011