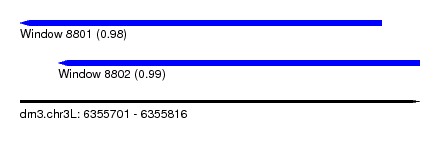
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,355,701 – 6,355,816 |
| Length | 115 |
| Max. P | 0.986584 |

| Location | 6,355,701 – 6,355,805 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 84.37 |
| Shannon entropy | 0.27655 |
| G+C content | 0.35516 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
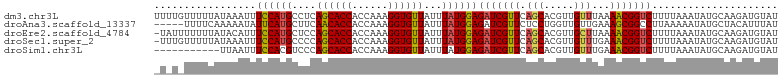
>dm3.chr3L 6355701 104 - 24543557 UUUUGUUUUUAUAAAUUUCCAUGCCUCAGCACCACCAAAGGUGUUAUUUAUGGAGAUCGUUCAGCACGUUGUUUAAAACGGUCUUUUAAAUAUGCAAGAUGUAU ((((((.((((......((((((....((((((......))))))...))))))(((((((.(((.....)))...)))))))...))))...))))))..... ( -24.10, z-score = -2.37, R) >droAna3.scaffold_13337 2708161 99 + 23293914 -----UUUUCAAAAAUAUUCAUGCUUCAACACCACCAAAGGUGUUAUUUAUGGAGAUCGUUCUCCUGGUUGUUGAAAGCGGCCUUAAAAAUAUGCUACAUUUAU -----.................((...((((((......))))))......(((((....))))).((((((.....))))))..........))......... ( -21.40, z-score = -1.77, R) >droEre2.scaffold_4784 9045224 103 - 25762168 -UAUUUUUUUAUACAUUUCCAUGCUCCAGCACCACCAAAGGUGUUAUUUAUGGAGAUCGUUCAGCACGUUGCUUAAAACGGUCUUUUAAAUAUGCAAGAUGUAU -.........((((((((...(((....(((((......)))))((((((.((((((((((.(((.....)))...)))))))))))))))).))))))))))) ( -28.30, z-score = -4.01, R) >droSec1.super_2 6320367 103 - 7591821 -UUUGUUUUUAUAAAUUUCCAUGCCCCAGCACCACCAAAGGUGUUAUUUAUGGAGAUCGUUCAGCACGUUGUUUGAAACGGUCUUUUAAAUAUGCAAGAUGUAU -(((((.((((......((((((....((((((......))))))...))))))(((((((((((.....).))).)))))))...))))...)))))...... ( -24.00, z-score = -2.00, R) >droSim1.chr3L 5854277 93 - 22553184 -----------UUAAUUUCCACGUCCCAGCACCACCAAAGGUGUUAUUUAUGGAGAUCGUUCAGCACGUUGUUUGAAACGGUCUUUUAAAUAUGCAAGAUGUAU -----------.........(((((...(((((......)))))((((((.((((((((((((((.....).))).)))))))))))))))).....))))).. ( -23.90, z-score = -2.54, R) >consensus _U_U_UUUUUAUAAAUUUCCAUGCCCCAGCACCACCAAAGGUGUUAUUUAUGGAGAUCGUUCAGCACGUUGUUUAAAACGGUCUUUUAAAUAUGCAAGAUGUAU .................((((((....((((((......))))))...))))))(((((((.(((.....)))...)))))))..................... (-18.56 = -18.76 + 0.20)
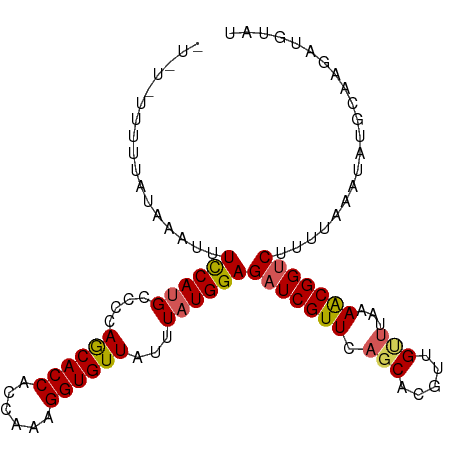
| Location | 6,355,712 – 6,355,816 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.40 |
| Shannon entropy | 0.34170 |
| G+C content | 0.34374 |
| Mean single sequence MFE | -21.51 |
| Consensus MFE | -17.29 |
| Energy contribution | -17.72 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6355712 104 - 24543557 CGUUUUUUUUUUUUUGUUUUUAUAAAUUUCCAUGCCUCAGCACCACCAAAGGUGUUAUUUAUGGAGAUCGUUCAGCACGUUGUUUAAAACGGUCUUUUAAAUAU ((((((........((((.......(((((((((....((((((......))))))...))))))))).....))))........))))))............. ( -20.59, z-score = -2.28, R) >droAna3.scaffold_13337 2708172 88 + 23293914 ----------------UUUUCAAAAAUAUUCAUGCUUCAACACCACCAAAGGUGUUAUUUAUGGAGAUCGUUCUCCUGGUUGUUGAAAGCGGCCUUAAAAAUAU ----------------......................((((((......))))))......(((((....))))).((((((.....)))))).......... ( -20.60, z-score = -2.23, R) >droSec1.super_2 6320378 103 - 7591821 -GGUUUUUUUUUUUUGUUUUUAUAAAUUUCCAUGCCCCAGCACCACCAAAGGUGUUAUUUAUGGAGAUCGUUCAGCACGUUGUUUGAAACGGUCUUUUAAAUAU -(((........(((((....))))).......)))..((((((......))))))(((((.((((((((((((((.....).))).))))))))))))))).. ( -22.16, z-score = -1.94, R) >droSim1.chr3L 5854288 93 - 22553184 -----------GGAUUUUUUUUUUAAUUUCCACGUCCCAGCACCACCAAAGGUGUUAUUUAUGGAGAUCGUUCAGCACGUUGUUUGAAACGGUCUUUUAAAUAU -----------(((..((......))..))).......((((((......))))))(((((.((((((((((((((.....).))).))))))))))))))).. ( -22.70, z-score = -2.69, R) >consensus ___________UUUUGUUUUUAUAAAUUUCCAUGCCCCAGCACCACCAAAGGUGUUAUUUAUGGAGAUCGUUCAGCACGUUGUUUAAAACGGUCUUUUAAAUAU ............................((((((....((((((......))))))...))))))(((((((.(((.....)))...))))))).......... (-17.29 = -17.72 + 0.44)

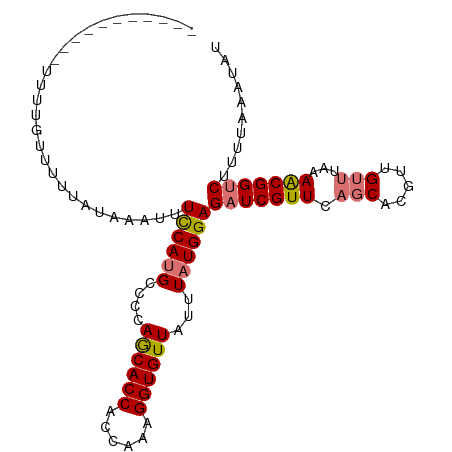
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:57 2011