| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,348,646 – 6,348,743 |
| Length | 97 |
| Max. P | 0.550421 |

| Location | 6,348,646 – 6,348,743 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 67.96 |
| Shannon entropy | 0.60197 |
| G+C content | 0.47552 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -9.74 |
| Energy contribution | -10.24 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.550421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
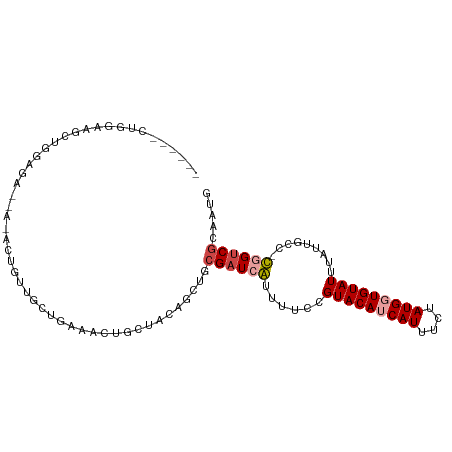
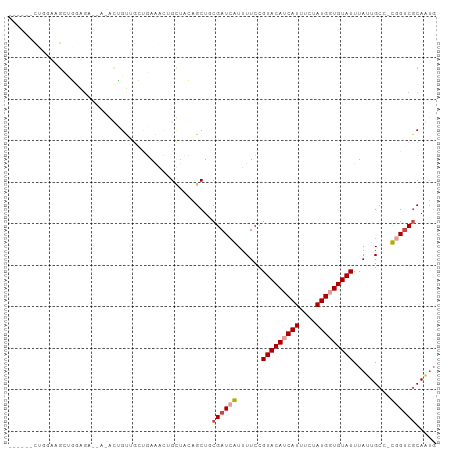
>dm3.chr3L 6348646 97 + 24543557 -GAGAGCUGGAAGCUGGUGA----GCUGUUGCUGAAGCUGCUUCAGCUCCGAUCGUUUUCCGUACAUCAUUUCUAUGCUGUAUUUAUUGCC-CGGUCGCAGUG -..((((((.((((.(((.(----((....)))...)))))))))))))((((((......(((((.(((....))).)))))........-))))))..... ( -29.74, z-score = -0.86, R) >droSim1.chr3L 5846873 95 + 22553184 ---GAGCUGGGAGCUGGAGA----GCUGUUGCUGAAGCUGCUUCAGCUCCGAUCGUUUUCCGUACAUCAUUUCUAUGCUGUAUUUAUUGCC-CGGUCGCAGUG ---..(((((((((((((((----(((........)))).))))))))))(((((......(((((.(((....))).)))))........-))))).)))). ( -37.54, z-score = -3.13, R) >droAna3.scaffold_13337 2701105 94 - 23293914 -----UCUUGAAGCUGAUGCUGAUGCUGAAGCUGAAGCUGCUUCAGCUGCGAUCGUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCC-CCGGCCCA--- -----....((((.((((((....((((((((.......)))))))).)).)))).)))).(((((((((....))))))))).....((.-...))...--- ( -29.90, z-score = -2.58, R) >droWil1.scaffold_180698 6350352 87 - 11422946 ---------------UAAGAC-AGAGAGUAAGAGAGACGGGGAGAGCUACGAUAAUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCCCUGGUCGCAAUG ---------------...(((-..((.((((........((((((..........))))))(((((((((....)))))))))...)))).)).)))...... ( -19.60, z-score = -0.61, R) >droGri2.scaffold_15110 15977486 102 - 24565398 GAAGGUUGGAUGGGUUGGGAUGAAAUGGCCGGCUAGAGAGUUAGAGCUGCGAUCAUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCC-UGGUCGCAAUG ....(((((((((((..(((..((((((((((((..........))))).).)))))))))(((((((((....))))))))).....)))-).))).)))). ( -26.90, z-score = -0.13, R) >droVir3.scaffold_13049 10665364 92 + 25233164 --------AACUGGCUGG--CUGGAUGGCUGGCUAGAGAGUUAGAGCUGCGAUCAUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCC-UGGUCGCAAUG --------..(((((..(--((....)))..)))))...........((((((((......(((((((((....)))))))))........-))))))))... ( -33.74, z-score = -2.99, R) >consensus ______CUGGAAGCUGGAGA__A_ACUGUUGCUGAAACUGCUACAGCUGCGAUCAUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCC_CGGUCGCAAUG .................................................((((((......(((((((((....))))))))).........))))))..... ( -9.74 = -10.24 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:56 2011