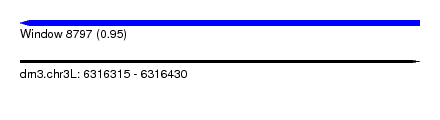
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,316,315 – 6,316,430 |
| Length | 115 |
| Max. P | 0.951241 |

| Location | 6,316,315 – 6,316,430 |
|---|---|
| Length | 115 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.87 |
| Shannon entropy | 0.57555 |
| G+C content | 0.53098 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -13.46 |
| Energy contribution | -13.16 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
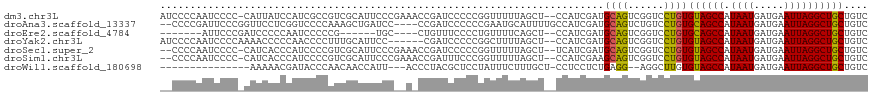
>dm3.chr3L 6316315 115 - 24543557 AUCCCCAAUCCCC-CAUUAUCCAUCGCCGUCGCAUUCCCGAAACCGAUCCCCCGGUUUUUAGCU--CCAUCGAUGCAGUCGGUCCUGUGUAGCCAUAAUGAUGAAUUAGGCUGCUGUC .............-.......((..((((.(((((((..(((((((......)))))))..)..--.....))))).).))))..)).((((((.((((.....)))))))))).... ( -26.91, z-score = -1.85, R) >droAna3.scaffold_13337 2671226 112 + 23293914 --CCCCGAUUCCCGGUUCCUCGGUCCCCAAAGCUGAUCC----CCGAUCCCCCCGAAUGCAUUUUGCCAUCGAUGCAGUCUGUCCUGUGCAGCCAUAAUGAUGAAUUAGGCUGCUGUC --....(((...(((....(((((.......)))))...----)))........((.((((((........)))))).)).)))....((((((.((((.....)))))))))).... ( -26.10, z-score = -0.74, R) >droEre2.scaffold_4784 9006643 99 - 25762168 -------AUUCCCGAUCCCCCAAUCCCCCG------UGC----CUGUUUCCCCUGUUUUCAGCU--CCAUCGAUGCAGUCGGUCCUGUGCAGCCAUAAUGAUGAAUUAGGCUGCUGUC -------....................(((------...----((((.((..(((....)))..--.....)).)))).)))......((((((.((((.....)))))))))).... ( -20.10, z-score = -0.91, R) >droYak2.chr3L 6902239 110 - 24197627 AUCCCCAAUCCCCAAAACCCCCAACCCCUUUGCAUUCC------CGAUCCCCCGGCUUUUAGCU--CCAUCGAUGCAGUCGGUCCUGUGUAGCCAUAAUGAUGAAUUAGGCUGCUGUC .........................((..((((((...------((((.....(((.....)))--..))))))))))..))......((((((.((((.....)))))))))).... ( -22.00, z-score = -1.28, R) >droSec1.super_2 6282806 113 - 7591821 --CCCCAAUCCCC-CAUCACCCAUCCCCGUCGCAUUCCCGAAACCGAUCCCCCGGUUUUUAGCU--UCAUCGAUGCAGUCGGUCCUGUGUAGCCAUAAUGAUGAAUUAGGCUGCUGUC --...........-............(((.(((((((..(((((((......)))))))..)..--.....))))).).)))......((((((.((((.....)))))))))).... ( -24.81, z-score = -1.50, R) >droSim1.chr3L 5815067 113 - 22553184 --CCCCAAUCCCC-CAUCACCCAUCCCCGUCGCAUUCCCGAAACCGAUUUCCCGGUUUUUAGCU--CCAUCGAAGCAGUCGGUCCUGUGUAGCCAUAAUGAUGAAUUAGGCUGCUGUC --...........-............(((.(........(((((((......)))))))..(((--.......))).).)))......((((((.((((.....)))))))))).... ( -24.20, z-score = -1.43, R) >droWil1.scaffold_180698 6316076 97 + 11422946 ---------------AAAAACGAUACCCAACAACCAUU---ACCCUACGCUCCUAUUUCUUUGCU-CCUCCUCUGAGG--AGGCUUGUGUAGCCAUAAUGAUGAAUUAGGCUGCUGUC ---------------......((((....((((((...---.......((............))(-((((....))))--))).))))((((((.((((.....)))))))))))))) ( -22.20, z-score = -2.01, R) >consensus __CCCCAAUCCCC_CAUCACCCAUCCCCGUCGCAUUCCC__A_CCGAUCCCCCGGUUUUUAGCU__CCAUCGAUGCAGUCGGUCCUGUGUAGCCAUAAUGAUGAAUUAGGCUGCUGUC ..........................................................................((((......))))((((((.((((.....)))))))))).... (-13.46 = -13.16 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:53 2011