| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,266,051 – 6,266,110 |
| Length | 59 |
| Max. P | 0.942432 |

| Location | 6,266,051 – 6,266,110 |
|---|---|
| Length | 59 |
| Sequences | 5 |
| Columns | 62 |
| Reading direction | forward |
| Mean pairwise identity | 61.03 |
| Shannon entropy | 0.66331 |
| G+C content | 0.47586 |
| Mean single sequence MFE | -11.67 |
| Consensus MFE | -6.56 |
| Energy contribution | -6.68 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.57 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
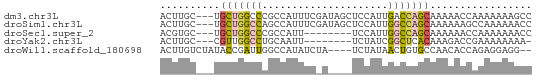
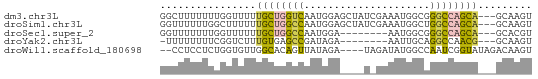
>dm3.chr3L 6266051 59 + 24543557 ACUUGC---UGCUGGCCCGCCAUUUCGAUAGCUCCAUUGACCAGCAAAAACCAAAAAAAGCC ..((((---((.(((....)))..(((((......))))).))))))............... ( -10.80, z-score = -0.93, R) >droSim1.chr3L 5759740 59 + 22553184 ACUUGC---UGCUGGCCAGCCAUUUCGAUAGCUCCAUUGGCCAGCAAAAAAGCCAAAAAACC .(((..---(((((((((((.((....)).)).....)))))))))...))).......... ( -16.90, z-score = -1.92, R) >droSec1.super_2 6231681 51 + 7591821 ACGUGC---UGCUGGCCCGCCAUU--------UCCAUUGGCCAGCAAAAAACCAAAAAAACC ......---((((((((.......--------......))))))))................ ( -14.12, z-score = -2.21, R) >droYak2.chr3L 6846878 50 + 24197627 ACUUGC---CGUUGGCCUGCAAUU--------UCUAUCGGCUCACAAAGACCGAAAAAAAA- ....((---(...)))........--------....((((.((.....)))))).......- ( -7.20, z-score = 0.08, R) >droWil1.scaffold_180698 6251611 56 - 11422946 ACUUGUCUAUACCGAUUGGCCAUAUCUA----UCUAUAACUGUGCCAACACCAGAGGAGG-- .(((.(((.......(((((((......----........)).)))))....))).))).-- ( -9.34, z-score = 0.09, R) >consensus ACUUGC___UGCUGGCCCGCCAUUUC_A____UCCAUUGGCCAGCAAAAACCAAAAAAAACC ..........(((((((.....................)))))))................. ( -6.56 = -6.68 + 0.12)
| Location | 6,266,051 – 6,266,110 |
|---|---|
| Length | 59 |
| Sequences | 5 |
| Columns | 62 |
| Reading direction | reverse |
| Mean pairwise identity | 61.03 |
| Shannon entropy | 0.66331 |
| G+C content | 0.47586 |
| Mean single sequence MFE | -15.52 |
| Consensus MFE | -8.10 |
| Energy contribution | -7.86 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.759476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
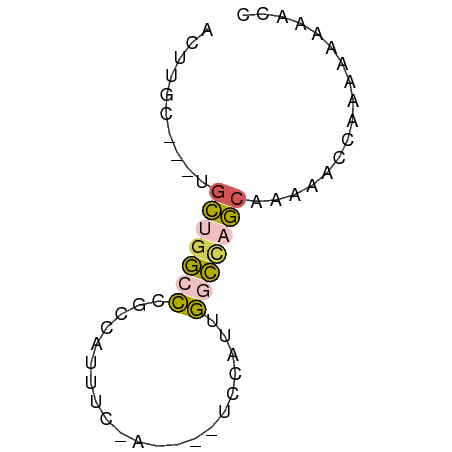
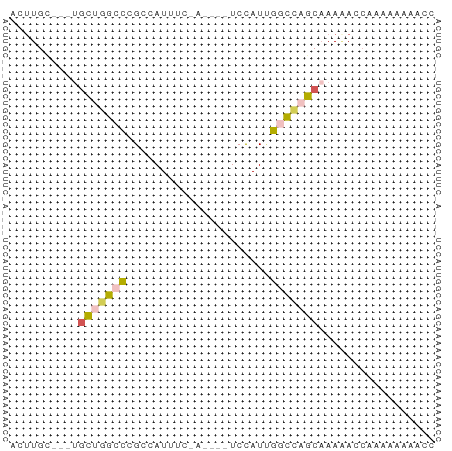
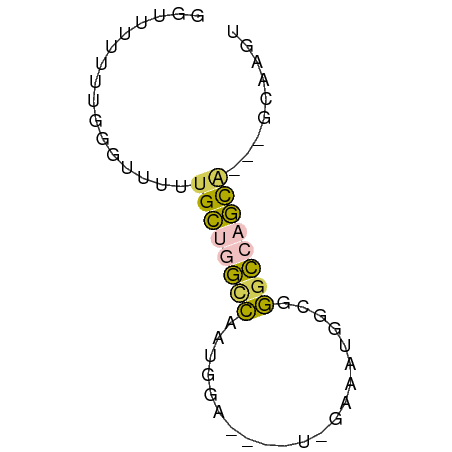
>dm3.chr3L 6266051 59 - 24543557 GGCUUUUUUUGGUUUUUGCUGGUCAAUGGAGCUAUCGAAAUGGCGGGCCAGCA---GCAAGU ((((......)))).(((((((((......(((((....))))).))))))))---)..... ( -17.60, z-score = -0.81, R) >droSim1.chr3L 5759740 59 - 22553184 GGUUUUUUGGCUUUUUUGCUGGCCAAUGGAGCUAUCGAAAUGGCUGGCCAGCA---GCAAGU .........((((..((((((((((.....(((((....))))))))))))))---).)))) ( -21.80, z-score = -1.57, R) >droSec1.super_2 6231681 51 - 7591821 GGUUUUUUUGGUUUUUUGCUGGCCAAUGGA--------AAUGGCGGGCCAGCA---GCACGU ...............(((((((((.((...--------.))....))))))))---)..... ( -17.10, z-score = -1.36, R) >droYak2.chr3L 6846878 50 - 24197627 -UUUUUUUUCGGUCUUUGUGAGCCGAUAGA--------AAUUGCAGGCCAACG---GCAAGU -.(((((.((((((.....)).)))).)))--------))((((.(.....).---)))).. ( -11.20, z-score = -0.52, R) >droWil1.scaffold_180698 6251611 56 + 11422946 --CCUCCUCUGGUGUUGGCACAGUUAUAGA----UAGAUAUGGCCAAUCGGUAUAGACAAGU --.((..((((..((((((.((.(((....----)))...)))))))).....))))..)). ( -9.90, z-score = 0.77, R) >consensus GGUUUUUUUGGGUUUUUGCUGGCCAAUGGA____U_GAAAUGGCGGGCCAGCA___GCAAGU .................(((((((.....................))))))).......... ( -8.10 = -7.86 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:50 2011