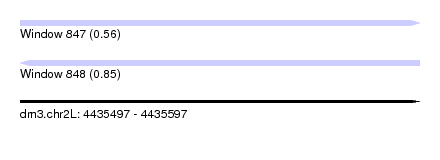
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,435,497 – 4,435,597 |
| Length | 100 |
| Max. P | 0.847068 |

| Location | 4,435,497 – 4,435,597 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.03 |
| Shannon entropy | 0.34084 |
| G+C content | 0.46545 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -15.61 |
| Energy contribution | -16.03 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.558073 |
| Prediction | RNA |
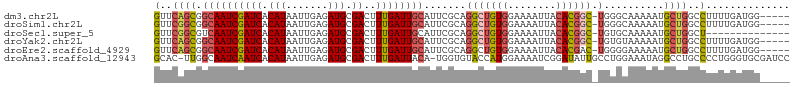
Download alignment: ClustalW | MAF
>dm3.chr2L 4435497 100 + 23011544 -----CCAUCAAAAGGCCAGCAUUUUUGCCCA-GCCGUGUAAUUUUCCACAGCCUGCGAAUGCAAUCAAAGUCGCAUCUCAAUUAUGUGAUCGAUUGCCGCUGAAC -----............((((...(((((...-((.(((........))).))..))))).((((((...(((((((.......))))))).)))))).))))... ( -26.40, z-score = -2.10, R) >droSim1.chr2L 4362569 100 + 22036055 -----CCAUCAAAAGGCCAGCAUUUUUGCCCA-GCCGUGUAAUUUUCCACAGCCUGCGAAUGCAAUCAAAGUCGCAUCUCAAUUAUGUGAUCGAUUGCCGCCGAAC -----.........(((.......(((((...-((.(((........))).))..))))).((((((...(((((((.......))))))).)))))).))).... ( -25.10, z-score = -1.88, R) >droSec1.super_5 2533565 91 + 5866729 --------------AGCCAGCAUUUUUGCACA-GCCGUGUAAUUUUCCACAGCCUGCGAAUGCAAUCAAAGUCGCAUCUCAAUUAUGUGAUCGAUUGACGCCGAAC --------------.....((...((((((..-((.(((........))).)).))))))..(((((...(((((((.......))))))).)))))..))..... ( -20.30, z-score = -0.51, R) >droYak2.chr2L 4469516 100 + 22324452 -----CCAUCAAAAGGCCAGCAUUUUUACACA-GCCGUGUAAUUUUCCACAGCCUGCGAAUGCAAUCAAAGUCGCAUCUCAAUUAUGUGAUCGAUUGCCGCUGAAC -----...(((..((((........((((((.-...)))))).........))))(((...((((((...(((((((.......))))))).)))))))))))).. ( -26.53, z-score = -2.53, R) >droEre2.scaffold_4929 4519819 100 + 26641161 -----CCAUCAAAAGGCCAGCAUUUUUCCCCA-GUCGUGUAAUUUUCCACAGCCUGCGAAUGCAAUCAAAGUCGCAUCUCAAUUAUGUGAUCGAUUGCCGCUGAAC -----............((((....(((..((-(.((((........))).).))).))).((((((...(((((((.......))))))).)))))).))))... ( -21.90, z-score = -1.37, R) >droAna3.scaffold_12943 1621636 104 - 5039921 GGAUCGCACCCAGGGGCAGGCCUAUUUCCAGGCAAUAUCCGAUUUUCCAUGGUACACCA-UGUAAUCAAAGUCGCAUCUCAAUUAUGUGAUUGAUUGCCAA-GUGC .....((((....(((....))).......((((((....((((...(((((....)))-)).))))..((((((((.......)))))))).))))))..-)))) ( -30.60, z-score = -1.58, R) >consensus _____CCAUCAAAAGGCCAGCAUUUUUGCACA_GCCGUGUAAUUUUCCACAGCCUGCGAAUGCAAUCAAAGUCGCAUCUCAAUUAUGUGAUCGAUUGCCGCUGAAC ..............(((..((((((.....((.((.(((........))).)).)).))))))((((...(((((((.......))))))).)))))))....... (-15.61 = -16.03 + 0.42)



| Location | 4,435,497 – 4,435,597 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.03 |
| Shannon entropy | 0.34084 |
| G+C content | 0.46545 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -20.01 |
| Energy contribution | -20.27 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4435497 100 - 23011544 GUUCAGCGGCAAUCGAUCACAUAAUUGAGAUGCGACUUUGAUUGCAUUCGCAGGCUGUGGAAAAUUACACGGC-UGGGCAAAAAUGCUGGCCUUUUGAUGG----- (..(((((((((((((((.(((.......))).))..))))))))....((.(((((((........))))))-)..)).....)))))..).........----- ( -33.20, z-score = -2.47, R) >droSim1.chr2L 4362569 100 - 22036055 GUUCGGCGGCAAUCGAUCACAUAAUUGAGAUGCGACUUUGAUUGCAUUCGCAGGCUGUGGAAAAUUACACGGC-UGGGCAAAAAUGCUGGCCUUUUGAUGG----- (..(((((((((((((((.(((.......))).))..))))))))....((.(((((((........))))))-)..)).....)))))..).........----- ( -32.50, z-score = -2.20, R) >droSec1.super_5 2533565 91 - 5866729 GUUCGGCGUCAAUCGAUCACAUAAUUGAGAUGCGACUUUGAUUGCAUUCGCAGGCUGUGGAAAAUUACACGGC-UGUGCAAAAAUGCUGGCU-------------- ..((((((.(((((((((.(((.......))).))..))))))))....((((((((((........))))))-).)))......)))))..-------------- ( -27.80, z-score = -1.47, R) >droYak2.chr2L 4469516 100 - 22324452 GUUCAGCGGCAAUCGAUCACAUAAUUGAGAUGCGACUUUGAUUGCAUUCGCAGGCUGUGGAAAAUUACACGGC-UGUGUAAAAAUGCUGGCCUUUUGAUGG----- (..(((((((((((((((.(((.......))).))..))))))))...(((((.(((((........))))))-))))......)))))..).........----- ( -31.90, z-score = -2.23, R) >droEre2.scaffold_4929 4519819 100 - 26641161 GUUCAGCGGCAAUCGAUCACAUAAUUGAGAUGCGACUUUGAUUGCAUUCGCAGGCUGUGGAAAAUUACACGAC-UGGGGAAAAAUGCUGGCCUUUUGAUGG----- (..(((((((((((((((.(((.......))).))..)))))))).(((.(((..((((........)))).)-)).)))....)))))..).........----- ( -29.30, z-score = -1.93, R) >droAna3.scaffold_12943 1621636 104 + 5039921 GCAC-UUGGCAAUCAAUCACAUAAUUGAGAUGCGACUUUGAUUACA-UGGUGUACCAUGGAAAAUCGGAUAUUGCCUGGAAAUAGGCCUGCCCCUGGGUGCGAUCC ((((-((((((.((.(((.((....)).)))..)).(((((((.((-(((....)))))...)))))))....(((((....))))).))))...))))))..... ( -30.80, z-score = -1.35, R) >consensus GUUCAGCGGCAAUCGAUCACAUAAUUGAGAUGCGACUUUGAUUGCAUUCGCAGGCUGUGGAAAAUUACACGGC_UGGGCAAAAAUGCUGGCCUUUUGAUGG_____ (..((((.((((((((((.(((.......))).))..)))))))).....((.((((((........)))))).)).........))))..).............. (-20.01 = -20.27 + 0.25)
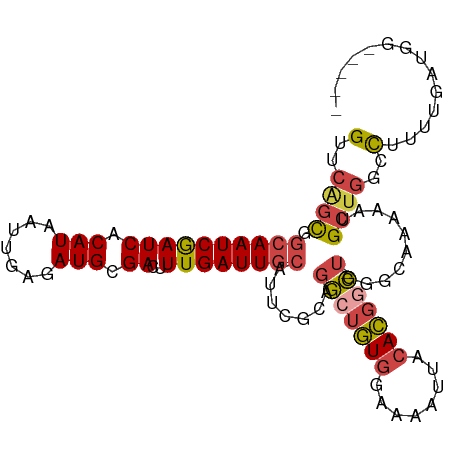
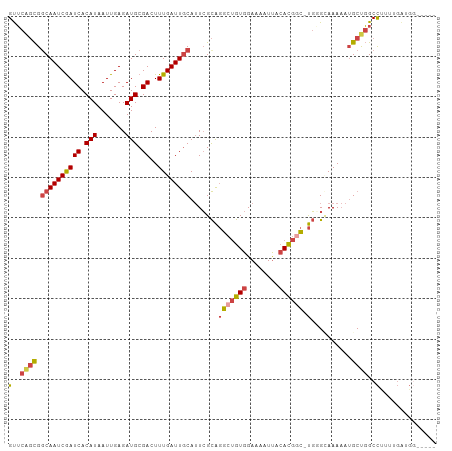
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:16:27 2011