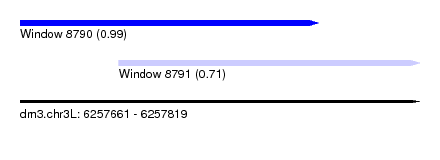
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,257,661 – 6,257,819 |
| Length | 158 |
| Max. P | 0.993324 |

| Location | 6,257,661 – 6,257,779 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.56 |
| Shannon entropy | 0.36470 |
| G+C content | 0.40405 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -19.95 |
| Energy contribution | -19.67 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
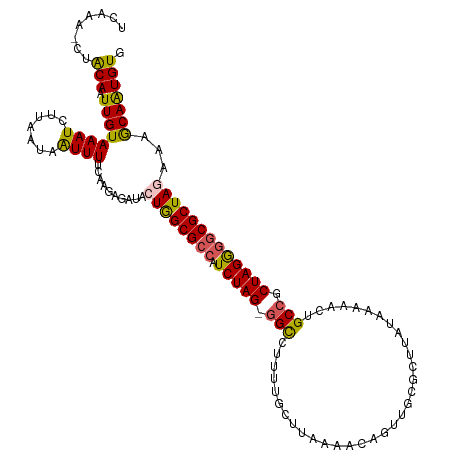
>dm3.chr3L 6257661 118 + 24543557 UCAAA-CUACAAUUGUAAAUCUGAAUAGUUUUCAAGAUUUACUGGCGCCAUCUAG-GGCCUUAUACUUGAGCCAGUUGCGCUUAUAAAAAAUGCCGCUAGGGGCGCUAAAAAGCAAUGUG .....-.((((...(((((((((((.....))).))))))))(((((((.(((((-(((........((((((....).)))))........))).))))))))))))........)))) ( -38.09, z-score = -3.40, R) >droEre2.scaffold_4784 8946846 119 + 25762168 UAAAAUCUACAAUUGUAAAUGUUCGUAAUUUUUAAAAGAUAGUGGCGGCCUCUAG-UGUCAUAAGCGAAAAUCAGUUGAGCUUAUAAAAACUGCCGCUAGGAGCGCUAAUAAGCAAUGUG .......((((.........((((...((((.....))))..(((((((....((-(...((((((..((.....))..))))))....)))))))))).))))(((....)))..)))) ( -23.50, z-score = -0.08, R) >droYak2.chr3L 6838064 119 + 24197627 UAAAAUCUGCAAUUGUAAAUUCUCAUCAUUUAUAAGAGAUACUGGCGCCCUCUAG-GGUCGCUUCCAUAAACCAGUUGCGCUUUCAAAAACUGCCGCUAGGGGCGCUAGAAAGCAGUGUG ......((((..((((((((.......))))))))......(((((((((.((((-((......))......(((((...........)))))...)))))))))))))...)))).... ( -36.30, z-score = -2.48, R) >droSec1.super_2 6223428 119 + 7591821 UCAAA-CUACAAUUGUAAAUCAUAAUAAUUUUCAAGAGAUACUAGCGCCAUCUAGCGGCCUUUUUCCUAAUACAGUUCCGCUUAUGAAAACUGCCGCUAGAGGCGCUAGAAAACAAUGUG .....-.((((.((((...............((....))..((((((((.(((((((((..(((((.(((...........))).)))))..)))))))))))))))))...)))))))) ( -38.30, z-score = -5.99, R) >droSim1.chr3L 5751474 119 + 22553184 UCAAA-CAACAAUUGUAAAUCUUAAUUAUUUUCAAGAGAUACUGGCGCCAUCUAGCGGCCUCUUGCUUAAUACAGUUGCGCCUUUGAUAACUGCCGCUAGGGGCGCUAGAAAACAAUGUG .....-..(((.((((..(((((............))))).((((((((.(((((((((..............(((((((....)).))))))))))))))))))))))...))))))). ( -36.63, z-score = -3.37, R) >consensus UCAAA_CUACAAUUGUAAAUCUUAAUAAUUUUCAAGAGAUACUGGCGCCAUCUAG_GGCCUUUUGCUUAAAACAGUUGCGCUUAUAAAAACUGCCGCUAGGGGCGCUAGAAAGCAAUGUG ........(((.((((((((.......))))..........((((((((.(((((.(((.................................))).)))))))))))))...))))))). (-19.95 = -19.67 + -0.28)

| Location | 6,257,700 – 6,257,819 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.95 |
| Shannon entropy | 0.37027 |
| G+C content | 0.44556 |
| Mean single sequence MFE | -36.74 |
| Consensus MFE | -21.59 |
| Energy contribution | -20.59 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.707029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
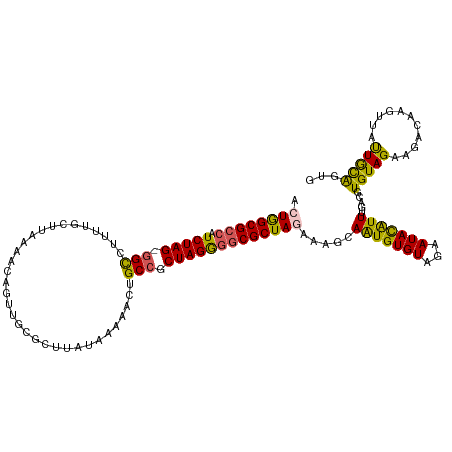
>dm3.chr3L 6257700 119 + 24543557 ACUGGCGCCAUCUAG-GGCCUUAUACUUGAGCCAGUUGCGCUUAUAAAAAAUGCCGCUAGGGGCGCUAAAAAGCAAUGUGUAGAAUACAUUUGACGUGUAGAAGACAAGUUAUUGCAGUG ..(((((((.(((((-(((........((((((....).)))))........))).))))))))))))....(((((((((....(((((.....)))))....)))...)))))).... ( -38.49, z-score = -2.02, R) >droEre2.scaffold_4784 8946886 119 + 25762168 AGUGGCGGCCUCUAG-UGUCAUAAGCGAAAAUCAGUUGAGCUUAUAAAAACUGCCGCUAGGAGCGCUAAUAAGCAAUGUGUAGAAUAUAUUUGACGUGUAGAAGAGAGGAGGCUAUGAUG .(((((..(((((..-..((((((((..((.....))..)))))).....(((((((.....))(((....)))...).)))).................))..)))))..))))).... ( -28.60, z-score = -0.19, R) >droYak2.chr3L 6838104 119 + 24197627 ACUGGCGCCCUCUAG-GGUCGCUUCCAUAAACCAGUUGCGCUUUCAAAAACUGCCGCUAGGGGCGCUAGAAAGCAGUGUGUAGAAUAUAUUUUACGUGUAGAGUAAAGUUGGCUAUAAUG .(((((((((.((((-((......))......(((((...........)))))...))))))))))))).........(((((.....(((((((.......)))))))...)))))... ( -36.10, z-score = -0.98, R) >droSec1.super_2 6223467 120 + 7591821 ACUAGCGCCAUCUAGCGGCCUUUUUCCUAAUACAGUUCCGCUUAUGAAAACUGCCGCUAGAGGCGCUAGAAAACAAUGUGUAGAAUACGUUUGACGUGUAGAAGACAAGUUAUUGCAGUA .((((((((.(((((((((..(((((.(((...........))).)))))..)))))))))))))))))....((((((((...(((((.....))))).....)))...)))))..... ( -42.20, z-score = -4.22, R) >droSim1.chr3L 5751513 120 + 22553184 ACUGGCGCCAUCUAGCGGCCUCUUGCUUAAUACAGUUGCGCCUUUGAUAACUGCCGCUAGGGGCGCUAGAAAACAAUGUGUAGAAUACAUUUGACGUGUAGAAGACAAGUUAUUGCAGUA .((((((((.(((((((((..............(((((((....)).))))))))))))))))))))))....((((((((....(((((.....)))))....)))...)))))..... ( -38.33, z-score = -1.56, R) >consensus ACUGGCGCCAUCUAG_GGCCUUUUGCUUAAAACAGUUGCGCUUAUAAAAACUGCCGCUAGGGGCGCUAGAAAGCAAUGUGUAGAAUACAUUUGACGUGUAGAAGACAAGUUAUUGCAGUG .((((((((.(((((.(((.................................))).))))))))))))).....(((((((...))))))).....(((((...........)))))... (-21.59 = -20.59 + -1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:48 2011