
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,178,195 – 6,178,300 |
| Length | 105 |
| Max. P | 0.976757 |

| Location | 6,178,195 – 6,178,298 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Shannon entropy | 0.32730 |
| G+C content | 0.46262 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -24.25 |
| Energy contribution | -25.61 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
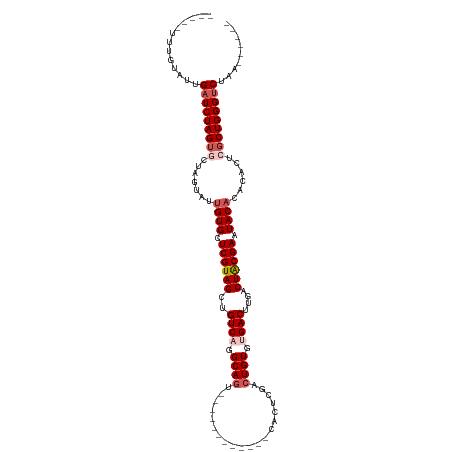
>dm3.chr3L 6178195 103 - 24543557 UGUAUUGUAUUGAUCUAGUGCUAGUAUUGUGCUCGUAGCUGUCCGGCAG----------UCUCACUCGACUGUGUGACUUGACUACGAAUACACACACUCGCUGGGUCUAAUA------ .......(((((((((((((..(((..((((.(((((((.(((..((((----------((......))))))..)))..).)))))).))))...)))))))))))).))))------ ( -35.30, z-score = -2.65, R) >droSim1.chr3L 5679553 96 - 22553184 -----UGUAUUGAUCUAGUGCUAGUAUUGUGCUCGUAGCUGUCAGGCAG------------UCACUCGACUGUGUGACUUGACUGCGAAUACACACACUCGCUGGGUCUAAUA------ -----..(((((((((((((..(((..((((.(((((((.((((.((((------------((....)))))).))))..).)))))).))))...)))))))))))).))))------ ( -39.70, z-score = -3.87, R) >droSec1.super_2 6122039 96 - 7591821 -----UGUAUUGAUCUAGUGCUAGUAUUGUGCUCGUAGCUGUCAGGCAG------------CCACUCGACUGUGUGACUUGACUGCGAAUACACACACUCGCUGGGUCUAAUA------ -----..(((((((((((((..(((..((((.(((((((.((((.((((------------........)))).))))..).)))))).))))...)))))))))))).))))------ ( -34.70, z-score = -2.24, R) >droYak2.chr3L 6767133 108 - 24197627 -----CGUUUUGAUCUAGUACUAUUAUUGUGCUCGUAGCUGUCACGCAAACAUAUUGACGCGAACUCGACUGUGCGACUUAACUACGAAUACACACACUCGCUGGGUCUAAUA------ -----......((((((((........((((.((((((..(((.((((..(.....)...((....))..)))).)))....)))))).)))).......)))))))).....------ ( -25.86, z-score = -1.03, R) >droEre2.scaffold_4784 8873711 114 - 25762168 -----CGUAUUGAUCUAGGACUAUUUUCGUGCUCGUAGCUGUCAGGCAGGCAUAUUUACGUGAACGCGACUGUGUGACUUAACUACGAAUACACACACUCGCUGGGCCUGGGUCUAAUA -----..((((((((((((.(((.....((.(.(((((.((((.....))))...))))).).))((((.((((((...............)))))).))))))).)))))))).)))) ( -34.06, z-score = -1.47, R) >consensus _____UGUAUUGAUCUAGUGCUAGUAUUGUGCUCGUAGCUGUCAGGCAG____________GCACUCGACUGUGUGACUUGACUACGAAUACACACACUCGCUGGGUCUAAUA______ ...........(((((((((.......((((.((((((..((((.((((....................)))).))))....)))))).))))......)))))))))........... (-24.25 = -25.61 + 1.36)


| Location | 6,178,197 – 6,178,300 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.67 |
| Shannon entropy | 0.32730 |
| G+C content | 0.46262 |
| Mean single sequence MFE | -33.84 |
| Consensus MFE | -25.23 |
| Energy contribution | -26.59 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6178197 103 - 24543557 UUUGUAUUGUAUUGAUCUAGUGCUAGUAUUGUGCUCGUAGCUGUCCGGCAGU----------CUCACUCGACUGUGUGACUUGACUACGAAUACACACACUCGCUGGGUCUAA------ .............(((((((((..(((..((((.(((((((.(((..(((((----------(......))))))..)))..).)))))).))))...))))))))))))...------ ( -35.20, z-score = -2.79, R) >droSim1.chr3L 5679555 96 - 22553184 -----UUUGUAUUGAUCUAGUGCUAGUAUUGUGCUCGUAGCUGUCAGGCAGU------------CACUCGACUGUGUGACUUGACUGCGAAUACACACACUCGCUGGGUCUAA------ -----........(((((((((..(((..((((.(((((((.((((.(((((------------(....)))))).))))..).)))))).))))...))))))))))))...------ ( -39.60, z-score = -3.98, R) >droSec1.super_2 6122041 96 - 7591821 -----UAUGUAUUGAUCUAGUGCUAGUAUUGUGCUCGUAGCUGUCAGGCAGC------------CACUCGACUGUGUGACUUGACUGCGAAUACACACACUCGCUGGGUCUAA------ -----........(((((((((..(((..((((.(((((((.((((.((((.------------.......)))).))))..).)))))).))))...))))))))))))...------ ( -34.60, z-score = -2.21, R) >droYak2.chr3L 6767135 108 - 24197627 -----UUCGUUUUGAUCUAGUACUAUUAUUGUGCUCGUAGCUGUCACGCAAACAUAUUGACGCGAACUCGACUGUGCGACUUAACUACGAAUACACACACUCGCUGGGUCUAA------ -----........((((((((........((((.((((((..(((.((((..(.....)...((....))..)))).)))....)))))).)))).......))))))))...------ ( -25.86, z-score = -1.03, R) >droEre2.scaffold_4784 8873713 114 - 25762168 -----UUCGUAUUGAUCUAGGACUAUUUUCGUGCUCGUAGCUGUCAGGCAGGCAUAUUUACGUGAACGCGACUGUGUGACUUAACUACGAAUACACACACUCGCUGGGCCUGGGUCUAA -----........((((((((.(((.....((.(.(((((.((((.....))))...))))).).))((((.((((((...............)))))).))))))).))))))))... ( -33.96, z-score = -1.41, R) >consensus _____UUUGUAUUGAUCUAGUGCUAGUAUUGUGCUCGUAGCUGUCAGGCAGU____________CACUCGACUGUGUGACUUGACUACGAAUACACACACUCGCUGGGUCUAA______ .............(((((((((.......((((.((((((..((((.((((....................)))).))))....)))))).))))......)))))))))......... (-25.23 = -26.59 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:43 2011