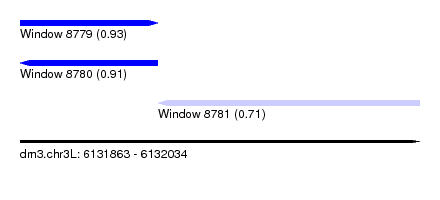
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,131,863 – 6,132,034 |
| Length | 171 |
| Max. P | 0.931172 |

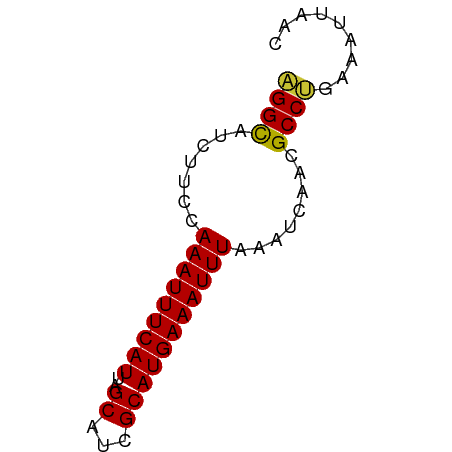
| Location | 6,131,863 – 6,131,922 |
|---|---|
| Length | 59 |
| Sequences | 8 |
| Columns | 59 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Shannon entropy | 0.08736 |
| G+C content | 0.34534 |
| Mean single sequence MFE | -9.92 |
| Consensus MFE | -9.06 |
| Energy contribution | -8.63 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6131863 59 + 24543557 AGGUAUCUUCCAAAUUUCAUUAGCAUCGCAUGAAAUUUAAAUCAACGCCUGAAAUUAAC ((((.......(((((((((..((...)))))))))))........))))......... ( -7.96, z-score = -1.49, R) >droSim1.chr3L 5632857 59 + 22553184 AGGUGUCUUCCAAAUUUCAUUAGCAUCGCAUGAAAUUUAAAUCAACGCCUGAAAUUAAC ((((((.....(((((((((..((...)))))))))))......))))))......... ( -11.90, z-score = -2.77, R) >droSec1.super_2 6076614 59 + 7591821 AGGUGUCUUCCAAAUUUCAUUAGCAUCGCAUGAAAUUUAAAUCAACGCCUGAAAUUAAC ((((((.....(((((((((..((...)))))))))))......))))))......... ( -11.90, z-score = -2.77, R) >droYak2.chr3L 6715810 59 + 24197627 AGGCAUCUUCCAAAUUUCAUUAGCAUCGCAUGAAAUUUAAAUCAACGCCUGAAAUUAAC ((((.......(((((((((..((...)))))))))))........))))......... ( -9.56, z-score = -1.97, R) >droEre2.scaffold_4784 8828339 59 + 25762168 AGGCAUCUUCCAAAUUUCAUUAGCAUCGCAUGAAAUUUAAAUCAACGCCUGAAAUUAAC ((((.......(((((((((..((...)))))))))))........))))......... ( -9.56, z-score = -1.97, R) >droAna3.scaffold_13337 2490586 59 - 23293914 AGGCAUCUUCCAAAUUUCAUUAGCAUCGCAUGAAAUUUAAAUCAACGCCAGAAAUUAAC .(((.......(((((((((..((...)))))))))))........))).......... ( -9.16, z-score = -2.27, R) >dp4.chrXR_group6 3776451 59 - 13314419 UGGCAUCUUCCAAAUUUCAUUAGCACCGCAUGAAAUUUAAAUCAGCGCCAGAAAUUAAC ((((.......(((((((((..((...)))))))))))........))))......... ( -9.66, z-score = -1.71, R) >droPer1.super_35 186814 59 + 973955 UGGCAUCUUCCAAAUUUCAUUAGCACCGCAUGAAAUUUAAAUCAGCGCCAGAAAUUAAC ((((.......(((((((((..((...)))))))))))........))))......... ( -9.66, z-score = -1.71, R) >consensus AGGCAUCUUCCAAAUUUCAUUAGCAUCGCAUGAAAUUUAAAUCAACGCCUGAAAUUAAC ((((.......(((((((((..((...)))))))))))........))))......... ( -9.06 = -8.63 + -0.42)

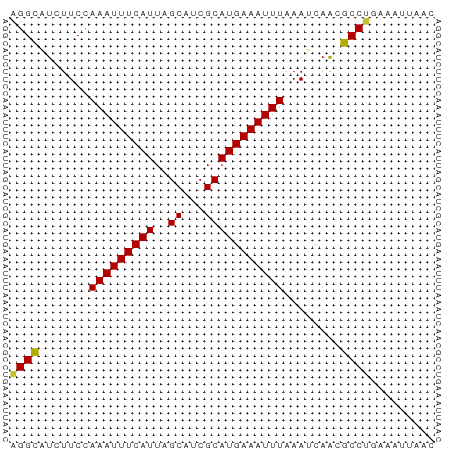
| Location | 6,131,863 – 6,131,922 |
|---|---|
| Length | 59 |
| Sequences | 8 |
| Columns | 59 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Shannon entropy | 0.08736 |
| G+C content | 0.34534 |
| Mean single sequence MFE | -15.75 |
| Consensus MFE | -12.07 |
| Energy contribution | -11.95 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6131863 59 - 24543557 GUUAAUUUCAGGCGUUGAUUUAAAUUUCAUGCGAUGCUAAUGAAAUUUGGAAGAUACCU .........(((..((..((((((((((((.........)))))))))))).))..))) ( -12.60, z-score = -1.89, R) >droSim1.chr3L 5632857 59 - 22553184 GUUAAUUUCAGGCGUUGAUUUAAAUUUCAUGCGAUGCUAAUGAAAUUUGGAAGACACCU .........(((.(((..((((((((((((.........)))))))))))).))).))) ( -14.70, z-score = -2.70, R) >droSec1.super_2 6076614 59 - 7591821 GUUAAUUUCAGGCGUUGAUUUAAAUUUCAUGCGAUGCUAAUGAAAUUUGGAAGACACCU .........(((.(((..((((((((((((.........)))))))))))).))).))) ( -14.70, z-score = -2.70, R) >droYak2.chr3L 6715810 59 - 24197627 GUUAAUUUCAGGCGUUGAUUUAAAUUUCAUGCGAUGCUAAUGAAAUUUGGAAGAUGCCU .........(((((((..((((((((((((.........)))))))))))).))))))) ( -18.30, z-score = -3.70, R) >droEre2.scaffold_4784 8828339 59 - 25762168 GUUAAUUUCAGGCGUUGAUUUAAAUUUCAUGCGAUGCUAAUGAAAUUUGGAAGAUGCCU .........(((((((..((((((((((((.........)))))))))))).))))))) ( -18.30, z-score = -3.70, R) >droAna3.scaffold_13337 2490586 59 + 23293914 GUUAAUUUCUGGCGUUGAUUUAAAUUUCAUGCGAUGCUAAUGAAAUUUGGAAGAUGCCU ..........((((((..((((((((((((.........)))))))))))).)))))). ( -17.60, z-score = -3.78, R) >dp4.chrXR_group6 3776451 59 + 13314419 GUUAAUUUCUGGCGCUGAUUUAAAUUUCAUGCGGUGCUAAUGAAAUUUGGAAGAUGCCA ..........((((((..((((((((((((.........)))))))))))))).)))). ( -14.90, z-score = -1.83, R) >droPer1.super_35 186814 59 - 973955 GUUAAUUUCUGGCGCUGAUUUAAAUUUCAUGCGGUGCUAAUGAAAUUUGGAAGAUGCCA ..........((((((..((((((((((((.........)))))))))))))).)))). ( -14.90, z-score = -1.83, R) >consensus GUUAAUUUCAGGCGUUGAUUUAAAUUUCAUGCGAUGCUAAUGAAAUUUGGAAGAUGCCU .........(((.(((..((((((((((((.........)))))))))))).))).))) (-12.07 = -11.95 + -0.12)

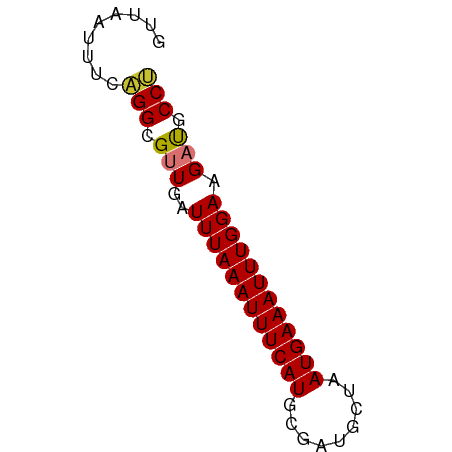
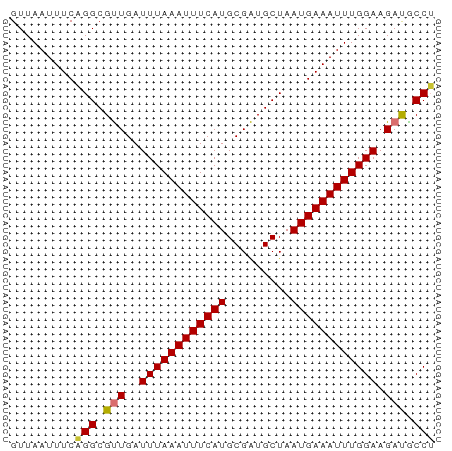
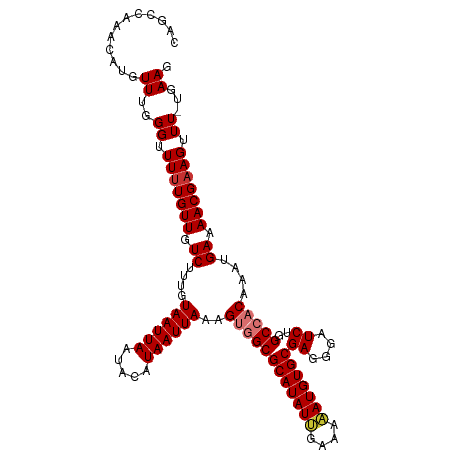
| Location | 6,131,922 – 6,132,034 |
|---|---|
| Length | 112 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 93.84 |
| Shannon entropy | 0.11309 |
| G+C content | 0.36301 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.706652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 6131922 112 - 24543557 CAGCCAAACAUGUUUGGGUUUUUGUUGUCUUUGUAAUUAAUACAUAAUUAAAGUGGCGCAUAUUGAAAAAUGUGCGAGGGAUCUGGCCACAAAUGAAAACGAAGUUU-UGAAG ...(((((....)))))(((((..((.......((((((.....))))))..((((((((((((....)))))))((....))..))))).))..))))).......-..... ( -29.80, z-score = -2.51, R) >droSim1.chr3L 5632916 112 - 22553184 CAGCCAAACAUGUUUGGGUUUUUGUUGUCUUUGUAAUUAAUACAUAAUUAAAGUGGCGCAUAUUGAAAAAUGUGCGAGGGAUCUGGCCACAAAUGAAAACGAAGUUU-UGAAG ...(((((....)))))(((((..((.......((((((.....))))))..((((((((((((....)))))))((....))..))))).))..))))).......-..... ( -29.80, z-score = -2.51, R) >droSec1.super_2 6076673 112 - 7591821 CAGCCAAACAUGUUUGGGUUUCUGUUGUCUUUGUAAUUAAUACAUAAUUAAAGUGGCGCAUAUUGAAAAAUGUGCGAGGGAUCUGGCCACAAAUGAAAACGAAGUUU-UGAAG ...(((((....)))))..(((......((((((..(((((.....))))).((((((((((((....)))))))((....))..)))))........))))))...-.))). ( -28.40, z-score = -1.97, R) >droYak2.chr3L 6715869 112 - 24197627 CAGCCAAACAUGUUUGGGUUUUUGUUGUCUUUGUAAUUAAUACAUAAUUAAAGUGGCGCAUAUUGAAAAAUGUGCGAGGGAUCUGGCCACAAAUGAAAACGAAGUUU-UGAAG ...(((((....)))))(((((..((.......((((((.....))))))..((((((((((((....)))))))((....))..))))).))..))))).......-..... ( -29.80, z-score = -2.51, R) >droEre2.scaffold_4784 8828398 112 - 25762168 CAGCCAAACAUGUUUGGGUUUUUGUUGUCUUUGUAAUUAAUACAUAAUUAAAGUGGCGCAUAUUGAAAAAUGUGCGAGGGAUCUGGCCACAAAUGAAAACGAAGUUU-UGAAG ...(((((....)))))(((((..((.......((((((.....))))))..((((((((((((....)))))))((....))..))))).))..))))).......-..... ( -29.80, z-score = -2.51, R) >droAna3.scaffold_13337 2490645 112 + 23293914 CGUUGAAACUUGUUUGGGUUUUUGUUGUCGUUGUAAUUAAUACAUAAUUAAAGUGGCGCAUAUUGAAAUAUGUGCGAGCGAUCUGGCCACGAAUGAAAACGAAGUUU-UGAAG ............((..((.(((((((.(((((.((((((.....))))))..(((((((((((......))))))((....))..))))).))))).))))))).))-..)). ( -27.30, z-score = -1.48, R) >dp4.chrXR_group6 3776510 113 + 13314419 CGUUGAAACUUGUUUGGGUUUUUGUUUUCUUUGUAAUUAAUACAUAAUUAAAGCCGCGCAUAUGGAAAUAUGUGCGAGCGAUCUGGCCACGAAUGAAAACGAAGUUUCUGAAG ............((..((.(((((((((((((((..(((((.....))))).(((((((((((....))))))))).)).........))))).))))))))))..))..)). ( -32.30, z-score = -3.09, R) >droPer1.super_35 186873 113 - 973955 CGUUGAAACUUGUUUGGGUUUUUGUUUUCUUUGUAAUUAAUACAUAAUUAAAGCCGCGCAUAUGGAAAUAUGUGCGAGCGAUCUGGCCACGAAUGAAAACGAAGUUUCUGAAG ............((..((.(((((((((((((((..(((((.....))))).(((((((((((....))))))))).)).........))))).))))))))))..))..)). ( -32.30, z-score = -3.09, R) >consensus CAGCCAAACAUGUUUGGGUUUUUGUUGUCUUUGUAAUUAAUACAUAAUUAAAGUGGCGCAUAUUGAAAAAUGUGCGAGGGAUCUGGCCACAAAUGAAAACGAAGUUU_UGAAG .................(((((((((.......((((((.....))))))..((((((((((((....)))))))((....))..))))).)))))))))............. (-21.42 = -21.86 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:39 2011