| Sequence ID | dm3.chr3L |
|---|---|
| Location | 6,100,821 – 6,100,913 |
| Length | 92 |
| Max. P | 0.732232 |

| Location | 6,100,821 – 6,100,913 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 70.94 |
| Shannon entropy | 0.55744 |
| G+C content | 0.39294 |
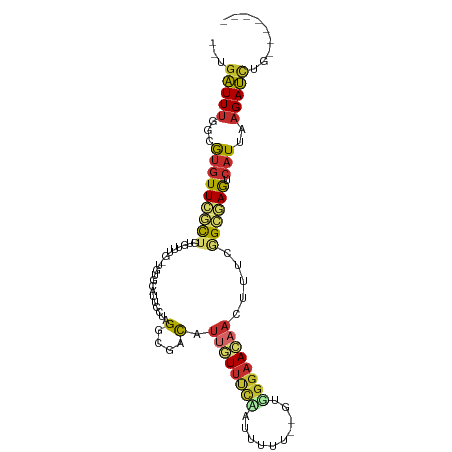
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -9.36 |
| Energy contribution | -9.80 |
| Covariance contribution | 0.44 |
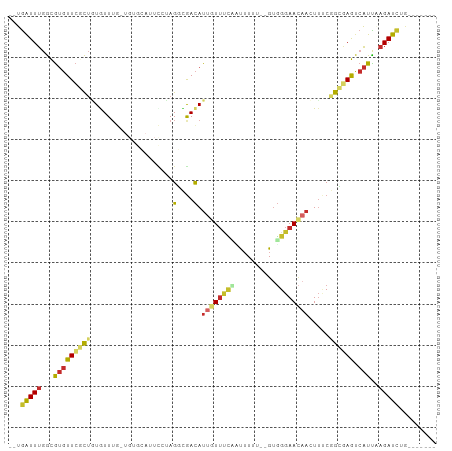
| Combinations/Pair | 1.61 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
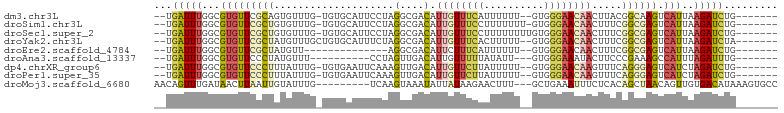
>dm3.chr3L 6100821 92 + 24543557 --UGAUUUGGCGUGUUCGCAGUGUUUG-UGUGCAUUCCUAGGCGACAUUGUUUCAUUUUUU--GUGGGAACAACUUACGGCAAGUCAUUAAGAUCUG------- --(((((((.(((((((((((((((.(-(.((......)).)))))))))).((((.....--)))))))).....))).)))))))..........------- ( -23.50, z-score = -0.77, R) >droSim1.chr3L 5604393 93 + 22553184 --UGAUUUGGCGUGUUCGCUGUGUUUG-UGUGCAUUCCUAGGCGACAUUGUUUCCUUUUUUU-GUGGGAACAACUUUCGGCGAGUCAUUAAGAUCUG------- --(((((((.(((((.((((..(..((-....))..)...)))))))((((((((.......-..))))))))....)).)))))))..........------- ( -25.90, z-score = -1.62, R) >droSec1.super_2 6049147 94 + 7591821 --UGAUUUGGCGUGUUCGCUGUGUUUG-UGUGCAUUCCUAGGCGACAUUGUUUCCUUUUUUUUGUGGGAACAACUUUCGGCGAGUCAUUAAGAUCUG------- --(((((((.(((((.((((..(..((-....))..)...)))))))((((((((..........))))))))....)).)))))))..........------- ( -25.80, z-score = -1.56, R) >droYak2.chr3L 6684193 93 + 24197627 --UGAUUUGGCGUGUUCGCUAUGUUUGCUGUGCAUUUCUAGGCGACAUUGUUUCACUUUUU--GUGGGAACAACUUUCGGCGAGUCAUUAAGAUCUA------- --(((((((.(((((.((((..(..(((...)))...)..)))))))(((((((((.....--)).)))))))....)).)))))))..........------- ( -23.10, z-score = -0.79, R) >droEre2.scaffold_4784 8799534 79 + 25762168 --UGAUUUGGCGUGUUCGCUAUGUU--------------AGGCGACAUUCUUUCAUUUUUU--GUGGGAACAACUUUCGGCGAGUCAUUAAGAUCUG------- --(((((((.(((((.((((.....--------------.)))))))((((..((.....)--)..)))).......)).)))))))..........------- ( -19.30, z-score = -1.24, R) >droAna3.scaffold_13337 2456645 82 - 23293914 --UGAUUUGGCGUGUUCCCUAUGUUU----------CCUAGUUGACAUUGUUUUUAUAUU---GUGGGAAAUACUUCCCGAAAGCCAUUUAGAUUUG------- --.....(((((((((..(((.....----------..)))..)))))............---.((((((....))))))...))))..........------- ( -17.20, z-score = -1.66, R) >dp4.chrXR_group6 3742000 92 - 13314419 --UGAUUUGGCGUGUUCCCUUUAUUUG-UGUGAAUUCAAAGUUGACAUUGUUCUUAUUUUU--GUGGGAACAAGUUUCAGGGAGUCAUCUAGAUCUG------- --.(((((((.(((((((((.......-(((.(((.....))).)))((((((((((....--)))))))))).....)))))).))))))))))..------- ( -28.20, z-score = -3.48, R) >droPer1.super_35 151852 92 + 973955 --UGAUUUGGCGUGUUCCCUUUAUUUG-UGUGAAUUCAAAGUUGACAUUGUUCUUAUUUUU--GUGGGAACAAGUUUCAGGGAGUCAUCUAGAUCUG------- --.(((((((.(((((((((.......-(((.(((.....))).)))((((((((((....--)))))))))).....)))))).))))))))))..------- ( -28.20, z-score = -3.48, R) >droMoj3.scaffold_6680 24158653 92 - 24764193 AACAGUUUGAUAACUUAAUUGUAUUUG---------UCAAGUAAAUAUUAUAAGAACUUU---GCUGAAAUUUCUCACAGCUAACAGUUGUGACAUAAAGUGCC ..((((.......((((...(((((((---------.....)))))))..))))......---)))).......(((((((.....)))))))........... ( -15.82, z-score = -0.38, R) >consensus __UGAUUUGGCGUGUUCGCUGUGUUUG_UGUGCAUUCCUAGGCGACAUUGUUUCAAUUUUU__GUGGGAACAACUUUCGGCGAGUCAUUAAGAUCUG_______ ...(((((...(((((((((....................(....).((((((((..........)))))))).....)))))).)))..)))))......... ( -9.36 = -9.80 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:06:33 2011